Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
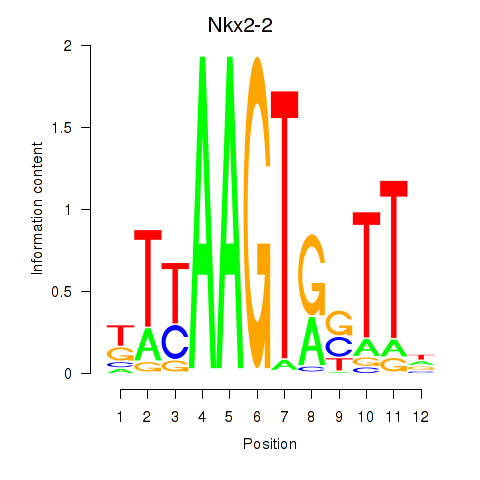
Results for Nkx2-2
Z-value: 1.27
Transcription factors associated with Nkx2-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-2
|
ENSMUSG00000027434.10 | NK2 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-2 | mm10_v2_chr2_-_147186389_147186413 | -0.13 | 4.6e-01 | Click! |
Activity profile of Nkx2-2 motif
Sorted Z-values of Nkx2-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_46039202 | 7.62 |
ENSMUST00000156200.1
|
Tmod1
|
tropomodulin 1 |
| chr10_+_88091070 | 6.76 |
ENSMUST00000048621.7
|
Pmch
|
pro-melanin-concentrating hormone |
| chr6_-_41704339 | 6.57 |
ENSMUST00000031899.8
|
Kel
|
Kell blood group |
| chr9_+_72438519 | 5.70 |
ENSMUST00000184604.1
|
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr9_+_72438534 | 5.36 |
ENSMUST00000034746.8
|
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chrX_+_9885622 | 5.04 |
ENSMUST00000067529.2
ENSMUST00000086165.3 |
Sytl5
|
synaptotagmin-like 5 |
| chr6_+_86078070 | 4.67 |
ENSMUST00000032069.5
|
Add2
|
adducin 2 (beta) |
| chr1_+_40439767 | 4.45 |
ENSMUST00000173514.1
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr1_+_40439627 | 4.26 |
ENSMUST00000097772.3
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr1_-_52953179 | 3.87 |
ENSMUST00000114492.1
|
1700019D03Rik
|
RIKEN cDNA 1700019D03 gene |
| chr13_-_19824234 | 3.49 |
ENSMUST00000065335.2
|
Gpr141
|
G protein-coupled receptor 141 |
| chr7_+_103915062 | 3.44 |
ENSMUST00000059011.3
ENSMUST00000167269.1 |
Olfr631
|
olfactory receptor 631 |
| chr6_+_5390387 | 3.36 |
ENSMUST00000183358.1
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr2_+_174450678 | 3.35 |
ENSMUST00000016399.5
|
Tubb1
|
tubulin, beta 1 class VI |
| chr7_+_110772604 | 3.29 |
ENSMUST00000005829.6
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr6_-_102464667 | 2.88 |
ENSMUST00000032159.6
|
Cntn3
|
contactin 3 |
| chr11_+_78301529 | 2.83 |
ENSMUST00000045026.3
|
Spag5
|
sperm associated antigen 5 |
| chr1_-_176807124 | 2.80 |
ENSMUST00000057037.7
|
Cep170
|
centrosomal protein 170 |
| chr15_-_75111684 | 2.75 |
ENSMUST00000100542.3
|
Ly6c2
|
lymphocyte antigen 6 complex, locus C2 |
| chrX_+_106027259 | 2.53 |
ENSMUST00000113557.1
|
Atp7a
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr2_-_140170528 | 2.50 |
ENSMUST00000046030.7
|
Esf1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr18_-_62179948 | 2.44 |
ENSMUST00000053640.3
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr6_-_70792155 | 2.34 |
ENSMUST00000066134.5
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr19_+_55180799 | 2.30 |
ENSMUST00000025936.5
|
Tectb
|
tectorin beta |
| chr14_+_53795455 | 2.19 |
ENSMUST00000103671.2
|
B230359F08Rik
|
RIKEN cDNA B230359F08 gene |
| chr1_-_65123108 | 2.12 |
ENSMUST00000050047.3
ENSMUST00000148020.1 |
D630023F18Rik
|
RIKEN cDNA D630023F18 gene |
| chr18_+_4993795 | 2.09 |
ENSMUST00000153016.1
|
Svil
|
supervillin |
| chr6_-_136857727 | 2.07 |
ENSMUST00000032341.2
|
Art4
|
ADP-ribosyltransferase 4 |
| chrX_+_106027300 | 2.04 |
ENSMUST00000055941.6
|
Atp7a
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr6_-_136401830 | 2.02 |
ENSMUST00000058713.7
|
E330021D16Rik
|
RIKEN cDNA E330021D16 gene |
| chr13_+_23756937 | 1.93 |
ENSMUST00000102965.2
|
Hist1h4b
|
histone cluster 1, H4b |
| chr15_+_66670749 | 1.92 |
ENSMUST00000065916.7
|
Tg
|
thyroglobulin |
| chr13_-_21753851 | 1.89 |
ENSMUST00000074752.2
|
Hist1h2ak
|
histone cluster 1, H2ak |
| chr7_-_100855403 | 1.88 |
ENSMUST00000156855.1
|
Relt
|
RELT tumor necrosis factor receptor |
| chr17_+_48346465 | 1.85 |
ENSMUST00000113237.3
|
Trem2
|
triggering receptor expressed on myeloid cells 2 |
| chr17_+_17887840 | 1.85 |
ENSMUST00000054871.5
ENSMUST00000064068.4 |
Fpr3
Fpr2
|
formyl peptide receptor 3 formyl peptide receptor 2 |
| chr9_-_96719404 | 1.81 |
ENSMUST00000140121.1
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr17_+_43568641 | 1.75 |
ENSMUST00000169694.1
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr17_+_43568475 | 1.75 |
ENSMUST00000167418.1
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chrX_+_164980592 | 1.73 |
ENSMUST00000101082.4
ENSMUST00000167446.1 ENSMUST00000057150.6 |
Fancb
|
Fanconi anemia, complementation group B |
| chr5_-_123140135 | 1.71 |
ENSMUST00000160099.1
|
AI480526
|
expressed sequence AI480526 |
| chr8_+_123332676 | 1.70 |
ENSMUST00000010298.6
|
Spire2
|
spire homolog 2 (Drosophila) |
| chr6_+_8520008 | 1.62 |
ENSMUST00000162567.1
ENSMUST00000161217.1 |
Glcci1
|
glucocorticoid induced transcript 1 |
| chr6_-_120357342 | 1.61 |
ENSMUST00000163827.1
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr3_+_108739658 | 1.58 |
ENSMUST00000133931.2
|
Aknad1
|
AKNA domain containing 1 |
| chr14_-_55788810 | 1.54 |
ENSMUST00000022830.6
ENSMUST00000168716.1 ENSMUST00000178399.1 |
Ripk3
|
receptor-interacting serine-threonine kinase 3 |
| chr7_-_43533171 | 1.52 |
ENSMUST00000004728.5
ENSMUST00000039861.5 |
Cd33
|
CD33 antigen |
| chr14_-_104522615 | 1.48 |
ENSMUST00000022716.2
|
Rnf219
|
ring finger protein 219 |
| chr19_-_5796924 | 1.47 |
ENSMUST00000174808.1
|
Malat1
|
metastasis associated lung adenocarcinoma transcript 1 (non-coding RNA) |
| chr9_+_51213683 | 1.45 |
ENSMUST00000034554.7
|
Pou2af1
|
POU domain, class 2, associating factor 1 |
| chr14_+_47663756 | 1.44 |
ENSMUST00000022391.7
|
Ktn1
|
kinectin 1 |
| chrX_+_142825698 | 1.44 |
ENSMUST00000112888.1
|
Tmem164
|
transmembrane protein 164 |
| chrX_+_101274023 | 1.43 |
ENSMUST00000117706.1
|
Med12
|
mediator of RNA polymerase II transcription, subunit 12 homolog (yeast) |
| chr2_+_52072823 | 1.41 |
ENSMUST00000112693.2
ENSMUST00000069794.5 |
Rif1
|
Rap1 interacting factor 1 homolog (yeast) |
| chr9_+_3017408 | 1.40 |
ENSMUST00000099049.3
|
Gm10719
|
predicted gene 10719 |
| chr6_-_56369625 | 1.40 |
ENSMUST00000170774.1
ENSMUST00000168944.1 ENSMUST00000166890.1 |
Pde1c
|
phosphodiesterase 1C |
| chr8_+_41239718 | 1.39 |
ENSMUST00000045218.7
|
Pcm1
|
pericentriolar material 1 |
| chr2_+_174076296 | 1.38 |
ENSMUST00000155000.1
ENSMUST00000134876.1 ENSMUST00000147038.1 |
Stx16
|
syntaxin 16 |
| chr13_-_8858762 | 1.37 |
ENSMUST00000176329.1
|
Wdr37
|
WD repeat domain 37 |
| chrX_+_75382384 | 1.36 |
ENSMUST00000033541.4
|
Fundc2
|
FUN14 domain containing 2 |
| chr13_+_104178797 | 1.36 |
ENSMUST00000022225.5
ENSMUST00000069187.5 |
Trim23
|
tripartite motif-containing 23 |
| chr5_+_10236829 | 1.33 |
ENSMUST00000101606.3
|
Gm10482
|
predicted gene 10482 |
| chr19_-_37178011 | 1.32 |
ENSMUST00000133988.1
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr12_+_55089202 | 1.32 |
ENSMUST00000021407.10
|
Srp54a
|
signal recognition particle 54A |
| chr6_-_120357440 | 1.30 |
ENSMUST00000112703.1
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr13_+_24614608 | 1.30 |
ENSMUST00000091694.3
|
Fam65b
|
family with sequence similarity 65, member B |
| chr2_+_83644435 | 1.29 |
ENSMUST00000081591.6
|
Zc3h15
|
zinc finger CCCH-type containing 15 |
| chr3_-_93015669 | 1.29 |
ENSMUST00000107301.1
ENSMUST00000029521.4 |
Crct1
|
cysteine-rich C-terminal 1 |
| chr2_+_62664279 | 1.29 |
ENSMUST00000028257.2
|
Gca
|
grancalcin |
| chr10_+_14523062 | 1.28 |
ENSMUST00000096020.5
|
Gm10335
|
predicted gene 10335 |
| chr8_-_24948771 | 1.23 |
ENSMUST00000119720.1
ENSMUST00000121438.2 |
Adam32
|
a disintegrin and metallopeptidase domain 32 |
| chr3_-_59220150 | 1.22 |
ENSMUST00000170388.1
|
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr6_-_120357422 | 1.22 |
ENSMUST00000032283.5
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr1_-_85736525 | 1.21 |
ENSMUST00000064788.7
|
A630001G21Rik
|
RIKEN cDNA A630001G21 gene |
| chr11_-_79523760 | 1.19 |
ENSMUST00000179322.1
|
Evi2b
|
ecotropic viral integration site 2b |
| chr18_+_82554463 | 1.19 |
ENSMUST00000062446.7
ENSMUST00000102812.4 ENSMUST00000075372.5 ENSMUST00000080658.4 ENSMUST00000152071.1 ENSMUST00000114674.3 ENSMUST00000142850.1 ENSMUST00000133193.1 ENSMUST00000123251.1 ENSMUST00000153478.1 ENSMUST00000132369.1 |
Mbp
|
myelin basic protein |
| chr9_+_3005125 | 1.19 |
ENSMUST00000179881.1
|
Gm11168
|
predicted gene 11168 |
| chr10_-_93891141 | 1.18 |
ENSMUST00000180840.1
|
Metap2
|
methionine aminopeptidase 2 |
| chr2_-_73453918 | 1.18 |
ENSMUST00000102679.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr15_-_10470490 | 1.12 |
ENSMUST00000136591.1
|
Dnajc21
|
DnaJ (Hsp40) homolog, subfamily C, member 21 |
| chr3_+_14533817 | 1.10 |
ENSMUST00000169079.1
ENSMUST00000091325.3 |
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr11_+_48800357 | 1.10 |
ENSMUST00000020640.7
|
Gnb2l1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 like 1 |
| chr5_+_110839973 | 1.10 |
ENSMUST00000066160.1
|
Chek2
|
checkpoint kinase 2 |
| chr15_-_73184840 | 1.09 |
ENSMUST00000044113.10
|
Ago2
|
argonaute RISC catalytic subunit 2 |
| chr6_+_29859686 | 1.08 |
ENSMUST00000134438.1
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr5_-_69592274 | 1.08 |
ENSMUST00000174233.1
ENSMUST00000120789.1 ENSMUST00000166298.1 |
Gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr5_-_30073554 | 1.08 |
ENSMUST00000026846.6
|
Tyms
|
thymidylate synthase |
| chr1_-_152386589 | 1.06 |
ENSMUST00000162371.1
|
Tsen15
|
tRNA splicing endonuclease 15 homolog (S. cerevisiae) |
| chr8_+_69300776 | 1.06 |
ENSMUST00000078257.6
|
D130040H23Rik
|
RIKEN cDNA D130040H23 gene |
| chr14_-_19418930 | 1.05 |
ENSMUST00000177817.1
|
Gm21738
|
predicted gene, 21738 |
| chr3_+_65528457 | 1.02 |
ENSMUST00000130705.1
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr3_+_5218516 | 1.02 |
ENSMUST00000175866.1
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr17_+_71019548 | 1.01 |
ENSMUST00000073211.5
ENSMUST00000179759.1 |
Myom1
|
myomesin 1 |
| chr19_-_40402267 | 0.99 |
ENSMUST00000099467.3
ENSMUST00000099466.3 ENSMUST00000165212.1 ENSMUST00000165469.1 |
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr11_-_26591729 | 0.99 |
ENSMUST00000109504.1
|
Vrk2
|
vaccinia related kinase 2 |
| chr14_-_54686060 | 0.99 |
ENSMUST00000125265.1
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr1_+_179961110 | 0.99 |
ENSMUST00000076687.5
ENSMUST00000097450.3 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr9_+_3034599 | 0.98 |
ENSMUST00000178641.1
|
Gm17535
|
predicted gene, 17535 |
| chr19_-_29753600 | 0.97 |
ENSMUST00000175764.1
|
9930021J03Rik
|
RIKEN cDNA 9930021J03 gene |
| chr14_-_72602945 | 0.97 |
ENSMUST00000162825.1
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr2_-_58160495 | 0.97 |
ENSMUST00000028175.6
|
Cytip
|
cytohesin 1 interacting protein |
| chr5_-_110839575 | 0.95 |
ENSMUST00000145318.1
|
Hscb
|
HscB iron-sulfur cluster co-chaperone homolog (E. coli) |
| chr3_-_130730375 | 0.95 |
ENSMUST00000079085.6
|
Rpl34
|
ribosomal protein L34 |
| chr8_+_123411424 | 0.94 |
ENSMUST00000071134.3
|
Tubb3
|
tubulin, beta 3 class III |
| chr10_+_57645834 | 0.94 |
ENSMUST00000177325.1
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr9_+_35559460 | 0.94 |
ENSMUST00000034615.3
ENSMUST00000121246.1 |
Pus3
|
pseudouridine synthase 3 |
| chr9_+_3025417 | 0.91 |
ENSMUST00000075573.6
|
Gm10717
|
predicted gene 10717 |
| chr11_+_3649759 | 0.90 |
ENSMUST00000140242.1
|
Morc2a
|
microrchidia 2A |
| chr13_+_38036989 | 0.87 |
ENSMUST00000021866.8
|
Riok1
|
RIO kinase 1 (yeast) |
| chr18_-_6241470 | 0.87 |
ENSMUST00000163210.1
|
Kif5b
|
kinesin family member 5B |
| chr10_+_57645861 | 0.86 |
ENSMUST00000177473.1
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr5_+_149439706 | 0.85 |
ENSMUST00000031667.4
|
Tex26
|
testis expressed 26 |
| chr2_+_69897255 | 0.84 |
ENSMUST00000131553.1
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr2_-_77946375 | 0.84 |
ENSMUST00000065889.3
|
Cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr2_-_25095125 | 0.84 |
ENSMUST00000114373.1
|
Noxa1
|
NADPH oxidase activator 1 |
| chr5_+_66968961 | 0.83 |
ENSMUST00000132991.1
|
Limch1
|
LIM and calponin homology domains 1 |
| chr2_-_77946331 | 0.83 |
ENSMUST00000111821.2
ENSMUST00000111818.1 |
Cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr9_+_3015654 | 0.82 |
ENSMUST00000099050.3
|
Gm10720
|
predicted gene 10720 |
| chr1_-_166002613 | 0.80 |
ENSMUST00000177358.1
ENSMUST00000160908.1 ENSMUST00000027850.8 ENSMUST00000160260.2 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr3_+_10366903 | 0.79 |
ENSMUST00000029049.5
|
Chmp4c
|
charged multivesicular body protein 4C |
| chr9_+_3018753 | 0.77 |
ENSMUST00000179272.1
|
Gm10719
|
predicted gene 10719 |
| chr2_+_43748802 | 0.76 |
ENSMUST00000112824.1
ENSMUST00000055776.7 |
Arhgap15
|
Rho GTPase activating protein 15 |
| chr2_-_25095149 | 0.75 |
ENSMUST00000044018.7
|
Noxa1
|
NADPH oxidase activator 1 |
| chr1_-_152386675 | 0.74 |
ENSMUST00000015124.8
|
Tsen15
|
tRNA splicing endonuclease 15 homolog (S. cerevisiae) |
| chr13_-_103764502 | 0.74 |
ENSMUST00000074616.5
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr17_-_65884902 | 0.74 |
ENSMUST00000024905.9
|
Ralbp1
|
ralA binding protein 1 |
| chr4_+_155891822 | 0.74 |
ENSMUST00000105584.3
ENSMUST00000079031.5 |
Acap3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr4_+_59626189 | 0.73 |
ENSMUST00000070150.4
ENSMUST00000052420.6 |
E130308A19Rik
|
RIKEN cDNA E130308A19 gene |
| chr4_+_126024506 | 0.73 |
ENSMUST00000106162.1
|
Csf3r
|
colony stimulating factor 3 receptor (granulocyte) |
| chrX_+_68821093 | 0.70 |
ENSMUST00000096420.2
|
Gm14698
|
predicted gene 14698 |
| chr12_+_38781093 | 0.69 |
ENSMUST00000161513.1
|
Etv1
|
ets variant gene 1 |
| chr12_+_107488632 | 0.68 |
ENSMUST00000082269.2
|
3110018I06Rik
|
RIKEN cDNA 3110018I06 gene |
| chr14_+_24490678 | 0.67 |
ENSMUST00000169826.1
ENSMUST00000112384.3 |
Rps24
|
ribosomal protein S24 |
| chr16_-_16527364 | 0.67 |
ENSMUST00000069284.7
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr4_-_129640959 | 0.65 |
ENSMUST00000132217.1
ENSMUST00000130017.1 ENSMUST00000154105.1 |
Txlna
|
taxilin alpha |
| chr4_-_45532470 | 0.64 |
ENSMUST00000147448.1
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr17_+_71616215 | 0.62 |
ENSMUST00000047086.9
|
Wdr43
|
WD repeat domain 43 |
| chr5_+_66968559 | 0.62 |
ENSMUST00000127184.1
|
Limch1
|
LIM and calponin homology domains 1 |
| chr14_-_73049107 | 0.62 |
ENSMUST00000044664.4
ENSMUST00000169168.1 |
Cysltr2
|
cysteinyl leukotriene receptor 2 |
| chr14_+_103513328 | 0.61 |
ENSMUST00000095576.3
|
Scel
|
sciellin |
| chr8_-_36953139 | 0.61 |
ENSMUST00000179501.1
|
Dlc1
|
deleted in liver cancer 1 |
| chr16_+_14580221 | 0.61 |
ENSMUST00000147024.1
|
A630010A05Rik
|
RIKEN cDNA A630010A05 gene |
| chr5_-_110839757 | 0.61 |
ENSMUST00000056937.5
|
Hscb
|
HscB iron-sulfur cluster co-chaperone homolog (E. coli) |
| chr3_+_65528404 | 0.59 |
ENSMUST00000047906.3
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr2_+_91650116 | 0.58 |
ENSMUST00000111331.2
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr17_+_78882003 | 0.57 |
ENSMUST00000180880.1
|
Gm26637
|
predicted gene, 26637 |
| chr3_+_3634145 | 0.56 |
ENSMUST00000108394.1
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr1_+_86064619 | 0.56 |
ENSMUST00000027432.8
|
Psmd1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr12_-_112860886 | 0.55 |
ENSMUST00000021729.7
|
Gpr132
|
G protein-coupled receptor 132 |
| chr11_-_69900930 | 0.54 |
ENSMUST00000018714.6
ENSMUST00000128046.1 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr5_+_87925579 | 0.54 |
ENSMUST00000001667.6
ENSMUST00000113267.1 |
Csn3
|
casein kappa |
| chr9_-_96719549 | 0.53 |
ENSMUST00000128269.1
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr17_-_32822200 | 0.52 |
ENSMUST00000179695.1
|
Zfp799
|
zinc finger protein 799 |
| chr18_+_12972225 | 0.51 |
ENSMUST00000025290.5
|
Impact
|
imprinted and ancient |
| chr13_-_38036923 | 0.50 |
ENSMUST00000110233.1
ENSMUST00000074969.4 ENSMUST00000131066.1 |
Cage1
|
cancer antigen 1 |
| chr1_-_135688094 | 0.50 |
ENSMUST00000112103.1
|
Nav1
|
neuron navigator 1 |
| chr1_+_179960472 | 0.50 |
ENSMUST00000097453.2
ENSMUST00000111117.1 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr3_+_138143888 | 0.50 |
ENSMUST00000161141.1
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr16_+_32496259 | 0.50 |
ENSMUST00000064192.7
|
Zdhhc19
|
zinc finger, DHHC domain containing 19 |
| chr12_-_83487708 | 0.50 |
ENSMUST00000177959.1
ENSMUST00000178756.1 |
Dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr11_-_69900949 | 0.49 |
ENSMUST00000102580.3
|
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr3_-_30140407 | 0.49 |
ENSMUST00000108271.3
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr6_+_88465409 | 0.49 |
ENSMUST00000032165.9
|
Ruvbl1
|
RuvB-like protein 1 |
| chr8_+_105880875 | 0.49 |
ENSMUST00000040254.9
ENSMUST00000119261.1 |
Edc4
|
enhancer of mRNA decapping 4 |
| chr11_-_71004387 | 0.48 |
ENSMUST00000124464.1
ENSMUST00000108527.1 |
Dhx33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chrM_+_11734 | 0.47 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr12_-_83984849 | 0.47 |
ENSMUST00000164935.2
|
Gm17673
|
predicted gene, 17673 |
| chr3_+_138143429 | 0.46 |
ENSMUST00000040321.6
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr7_-_123369870 | 0.45 |
ENSMUST00000106442.2
ENSMUST00000098060.3 ENSMUST00000167309.1 |
Arhgap17
|
Rho GTPase activating protein 17 |
| chr10_-_13388830 | 0.45 |
ENSMUST00000079698.5
|
Phactr2
|
phosphatase and actin regulator 2 |
| chrX_+_107149580 | 0.45 |
ENSMUST00000137107.1
ENSMUST00000067249.2 |
A630033H20Rik
|
RIKEN cDNA A630033H20 gene |
| chr19_+_8819401 | 0.45 |
ENSMUST00000096753.3
|
Hnrnpul2
|
heterogeneous nuclear ribonucleoprotein U-like 2 |
| chr6_+_143285988 | 0.45 |
ENSMUST00000160951.1
|
D6Ertd474e
|
DNA segment, Chr 6, ERATO Doi 474, expressed |
| chr2_+_163661495 | 0.44 |
ENSMUST00000135537.1
|
Pkig
|
protein kinase inhibitor, gamma |
| chr19_+_34100943 | 0.44 |
ENSMUST00000025685.6
|
Lipm
|
lipase, family member M |
| chr12_+_73901370 | 0.43 |
ENSMUST00000110461.1
|
Hif1a
|
hypoxia inducible factor 1, alpha subunit |
| chr11_+_66956620 | 0.42 |
ENSMUST00000150220.1
|
9130409J20Rik
|
RIKEN cDNA 9130409J20 gene |
| chr15_-_102350692 | 0.41 |
ENSMUST00000041208.7
|
Aaas
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr2_+_3704787 | 0.41 |
ENSMUST00000115054.2
|
Fam107b
|
family with sequence similarity 107, member B |
| chr18_-_6241486 | 0.41 |
ENSMUST00000025083.7
|
Kif5b
|
kinesin family member 5B |
| chr3_-_92687209 | 0.40 |
ENSMUST00000029524.3
|
Lce1d
|
late cornified envelope 1D |
| chr4_-_43010226 | 0.39 |
ENSMUST00000030165.4
|
Fancg
|
Fanconi anemia, complementation group G |
| chr3_+_32515295 | 0.38 |
ENSMUST00000029203.7
|
Zfp639
|
zinc finger protein 639 |
| chr10_-_116896879 | 0.38 |
ENSMUST00000048229.7
|
Myrfl
|
myelin regulatory factor-like |
| chrX_+_6415736 | 0.38 |
ENSMUST00000143641.3
|
Shroom4
|
shroom family member 4 |
| chr1_-_9748376 | 0.38 |
ENSMUST00000057438.6
|
Vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chrX_+_107149454 | 0.37 |
ENSMUST00000125676.1
ENSMUST00000180182.1 |
A630033H20Rik
|
RIKEN cDNA A630033H20 gene |
| chr11_-_69900886 | 0.37 |
ENSMUST00000108621.2
ENSMUST00000100969.2 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr6_+_122990367 | 0.36 |
ENSMUST00000079379.2
|
Clec4a4
|
C-type lectin domain family 4, member a4 |
| chr5_-_3596071 | 0.36 |
ENSMUST00000121877.1
|
Rbm48
|
RNA binding motif protein 48 |
| chr19_+_58759700 | 0.36 |
ENSMUST00000026081.3
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr6_-_23650206 | 0.35 |
ENSMUST00000115354.1
|
Rnf133
|
ring finger protein 133 |
| chr8_+_40354303 | 0.35 |
ENSMUST00000136835.1
|
Micu3
|
mitochondrial calcium uptake family, member 3 |
| chr2_+_71873224 | 0.35 |
ENSMUST00000006669.5
|
Pdk1
|
pyruvate dehydrogenase kinase, isoenzyme 1 |
| chr3_+_138143483 | 0.35 |
ENSMUST00000162864.1
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr11_+_99785191 | 0.35 |
ENSMUST00000105059.2
|
Krtap4-9
|
keratin associated protein 4-9 |
| chr3_+_138143846 | 0.34 |
ENSMUST00000159481.1
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr2_+_91650169 | 0.34 |
ENSMUST00000090614.4
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr9_-_4796218 | 0.33 |
ENSMUST00000027020.6
ENSMUST00000063508.7 ENSMUST00000163309.1 |
Gria4
|
glutamate receptor, ionotropic, AMPA4 (alpha 4) |
| chr3_-_106024911 | 0.33 |
ENSMUST00000066537.3
ENSMUST00000054973.3 |
Chi3l7
|
chitinase 3-like 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 1.5 | 4.6 | GO:1904732 | elastin biosynthetic process(GO:0051542) regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 1.3 | 7.6 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 1.1 | 6.6 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.8 | 8.7 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.7 | 2.8 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.6 | 1.9 | GO:0002582 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.6 | 2.4 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.6 | 2.3 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.6 | 3.5 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.6 | 11.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.6 | 1.7 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.5 | 1.4 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.4 | 1.3 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.4 | 1.7 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.4 | 1.2 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.4 | 1.5 | GO:2000449 | regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) |
| 0.4 | 3.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.4 | 1.1 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 0.3 | 3.4 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.3 | 1.2 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.3 | 1.5 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.3 | 1.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.3 | 1.1 | GO:0046073 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 0.3 | 0.8 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.3 | 1.3 | GO:0072383 | stress granule disassembly(GO:0035617) plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 1.5 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.2 | 1.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 1.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 0.8 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 1.4 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 1.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 | 1.9 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 4.7 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.2 | 0.5 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.2 | 0.9 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 0.9 | GO:0032439 | endosome localization(GO:0032439) negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 0.4 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 1.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 1.8 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 2.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.7 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 5.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 5.1 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.8 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 1.0 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.1 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 1.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 1.9 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 1.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 1.6 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.1 | 1.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 1.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 1.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.2 | GO:0071963 | unidimensional cell growth(GO:0009826) establishment or maintenance of cell polarity regulating cell shape(GO:0071963) regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 | 0.4 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.7 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.2 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 0.3 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 | 1.8 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.5 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.1 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 0.5 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 1.1 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.1 | 1.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.5 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.7 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 1.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.2 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.3 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 3.3 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 1.0 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 1.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.6 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.4 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 1.0 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.4 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.8 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.5 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.2 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.9 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 1.0 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 3.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 2.7 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.3 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.6 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.1 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.0 | 0.3 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 2.2 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.0 | 0.1 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.9 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.5 | 3.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 1.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 1.5 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.2 | 2.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 0.9 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 1.4 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.2 | 1.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.2 | 3.5 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.2 | 7.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 2.8 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 1.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 1.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 4.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 1.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 1.7 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.1 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 1.0 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 11.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 0.3 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.8 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 2.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.6 | GO:0031597 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 8.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 1.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 2.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 8.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 1.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 2.0 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.2 | GO:0035371 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.0 | 3.5 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 4.4 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 1.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 5.2 | GO:0005813 | centrosome(GO:0005813) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.7 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 1.1 | 4.6 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.6 | 2.4 | GO:0031711 | beta-adrenergic receptor activity(GO:0004939) bradykinin receptor binding(GO:0031711) |
| 0.4 | 7.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 3.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 3.5 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.3 | 3.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.3 | 1.5 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.3 | 1.8 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.3 | 1.8 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.3 | 1.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.2 | 1.8 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.2 | 1.4 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 1.3 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.2 | 1.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 5.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 1.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 1.9 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.2 | 0.6 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 3.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 2.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 2.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 1.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 2.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 2.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 5.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 4.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 1.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 5.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 6.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 2.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.5 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 1.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.2 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 3.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 3.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 2.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 4.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 2.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 2.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 2.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.6 | PID ENDOTHELIN PATHWAY | Endothelins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 3.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 3.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 7.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.1 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 4.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 1.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 2.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.6 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 7.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.0 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 3.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 3.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 1.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.4 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.4 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |