Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
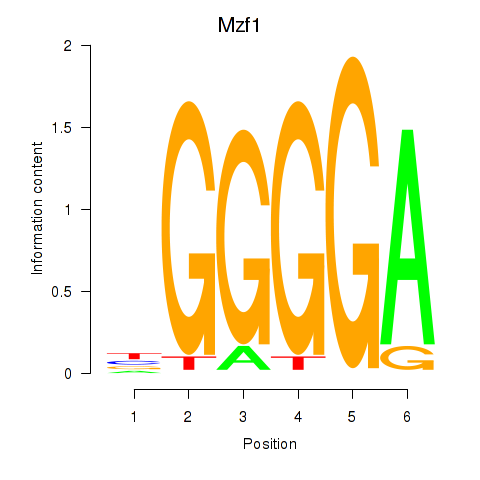
Results for Mzf1
Z-value: 1.26
Transcription factors associated with Mzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mzf1
|
ENSMUSG00000030380.10 | myeloid zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mzf1 | mm10_v2_chr7_-_13054665_13054764 | -0.21 | 2.2e-01 | Click! |
Activity profile of Mzf1 motif
Sorted Z-values of Mzf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_60582152 | 2.40 |
ENSMUST00000098047.2
|
Mup10
|
major urinary protein 10 |
| chr17_+_29490812 | 2.20 |
ENSMUST00000024811.6
|
Pim1
|
proviral integration site 1 |
| chr4_-_60222580 | 2.20 |
ENSMUST00000095058.4
ENSMUST00000163931.1 |
Mup8
|
major urinary protein 8 |
| chr4_-_61303998 | 2.19 |
ENSMUST00000071005.8
ENSMUST00000075206.5 |
Mup14
|
major urinary protein 14 |
| chr4_-_60421933 | 2.09 |
ENSMUST00000107506.2
ENSMUST00000122381.1 ENSMUST00000118759.1 ENSMUST00000122177.1 |
Mup9
|
major urinary protein 9 |
| chr4_-_61303802 | 2.05 |
ENSMUST00000125461.1
|
Mup14
|
major urinary protein 14 |
| chr4_-_60741275 | 2.05 |
ENSMUST00000117932.1
|
Mup12
|
major urinary protein 12 |
| chr4_-_60501903 | 2.02 |
ENSMUST00000084548.4
ENSMUST00000103012.3 ENSMUST00000107499.3 |
Mup1
|
major urinary protein 1 |
| chr14_-_54712139 | 1.95 |
ENSMUST00000064290.6
|
Cebpe
|
CCAAT/enhancer binding protein (C/EBP), epsilon |
| chr9_-_114564315 | 1.71 |
ENSMUST00000111816.2
|
Trim71
|
tripartite motif-containing 71 |
| chr7_+_44384604 | 1.57 |
ENSMUST00000130707.1
ENSMUST00000130844.1 |
Syt3
|
synaptotagmin III |
| chr10_-_114801364 | 1.48 |
ENSMUST00000061632.7
|
Trhde
|
TRH-degrading enzyme |
| chr13_+_91461050 | 1.28 |
ENSMUST00000004094.8
ENSMUST00000042122.8 |
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr4_-_62150810 | 1.28 |
ENSMUST00000077719.3
|
Mup21
|
major urinary protein 21 |
| chr11_+_32276893 | 1.26 |
ENSMUST00000145569.1
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr14_-_69284982 | 1.18 |
ENSMUST00000183882.1
ENSMUST00000037064.4 |
Slc25a37
|
solute carrier family 25, member 37 |
| chr19_+_44333092 | 1.17 |
ENSMUST00000058856.8
|
Scd4
|
stearoyl-coenzyme A desaturase 4 |
| chr7_+_126776939 | 1.10 |
ENSMUST00000038614.5
ENSMUST00000170882.1 ENSMUST00000106359.1 ENSMUST00000106357.1 ENSMUST00000145762.1 ENSMUST00000132643.1 ENSMUST00000106356.1 |
Ypel3
|
yippee-like 3 (Drosophila) |
| chr18_+_61687911 | 1.08 |
ENSMUST00000025471.2
|
Il17b
|
interleukin 17B |
| chr11_+_32276400 | 1.02 |
ENSMUST00000020531.2
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr7_+_44384803 | 1.01 |
ENSMUST00000120262.1
|
Syt3
|
synaptotagmin III |
| chr4_-_106464167 | 1.01 |
ENSMUST00000049507.5
|
Pcsk9
|
proprotein convertase subtilisin/kexin type 9 |
| chr7_+_126781483 | 0.95 |
ENSMUST00000172352.1
ENSMUST00000094037.4 |
Tbx6
|
T-box 6 |
| chr17_+_3532554 | 0.91 |
ENSMUST00000168560.1
|
Cldn20
|
claudin 20 |
| chr5_-_24601961 | 0.89 |
ENSMUST00000030791.7
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr4_+_117849361 | 0.87 |
ENSMUST00000163288.1
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr4_-_59960659 | 0.87 |
ENSMUST00000075973.2
|
Mup4
|
major urinary protein 4 |
| chr4_+_127172866 | 0.82 |
ENSMUST00000106094.2
|
Dlgap3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr4_+_117849193 | 0.81 |
ENSMUST00000132043.2
ENSMUST00000169990.1 |
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr11_+_96282648 | 0.78 |
ENSMUST00000168043.1
|
Hoxb8
|
homeobox B8 |
| chr13_-_76056996 | 0.77 |
ENSMUST00000056130.4
|
Gpr150
|
G protein-coupled receptor 150 |
| chr11_+_23256566 | 0.75 |
ENSMUST00000136235.1
|
Xpo1
|
exportin 1, CRM1 homolog (yeast) |
| chr5_-_24392012 | 0.75 |
ENSMUST00000059401.6
|
Atg9b
|
autophagy related 9B |
| chr6_+_39420378 | 0.72 |
ENSMUST00000090237.2
|
Gm10244
|
predicted gene 10244 |
| chr1_+_75400070 | 0.72 |
ENSMUST00000113589.1
|
Speg
|
SPEG complex locus |
| chr1_-_173333503 | 0.72 |
ENSMUST00000038227.4
|
Darc
|
Duffy blood group, chemokine receptor |
| chrX_-_143933089 | 0.71 |
ENSMUST00000087313.3
|
Dcx
|
doublecortin |
| chr11_-_97150025 | 0.70 |
ENSMUST00000118375.1
|
Tbkbp1
|
TBK1 binding protein 1 |
| chrX_-_7964166 | 0.69 |
ENSMUST00000128449.1
|
Gata1
|
GATA binding protein 1 |
| chr6_-_39420418 | 0.67 |
ENSMUST00000031985.6
|
Mkrn1
|
makorin, ring finger protein, 1 |
| chr11_+_24080664 | 0.67 |
ENSMUST00000118955.1
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
| chr15_-_76669811 | 0.67 |
ENSMUST00000037824.4
|
Foxh1
|
forkhead box H1 |
| chr11_-_95514570 | 0.66 |
ENSMUST00000058866.7
|
Nxph3
|
neurexophilin 3 |
| chr2_-_153241402 | 0.66 |
ENSMUST00000056924.7
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr4_-_109476666 | 0.64 |
ENSMUST00000030284.3
|
Rnf11
|
ring finger protein 11 |
| chr7_+_30712209 | 0.63 |
ENSMUST00000005692.6
ENSMUST00000170371.1 |
Atp4a
|
ATPase, H+/K+ exchanging, gastric, alpha polypeptide |
| chrX_+_101254528 | 0.63 |
ENSMUST00000062000.4
|
Foxo4
|
forkhead box O4 |
| chrX_-_49797700 | 0.63 |
ENSMUST00000033442.7
ENSMUST00000114891.1 |
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr16_+_13986596 | 0.62 |
ENSMUST00000056521.5
ENSMUST00000118412.1 ENSMUST00000131608.1 |
2900011O08Rik
|
RIKEN cDNA 2900011O08 gene |
| chr2_+_153492790 | 0.62 |
ENSMUST00000109783.1
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chrX_-_47892396 | 0.62 |
ENSMUST00000153548.2
|
Smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr3_+_107895916 | 0.61 |
ENSMUST00000172247.1
ENSMUST00000167387.1 |
Gstm5
|
glutathione S-transferase, mu 5 |
| chr16_+_44173271 | 0.61 |
ENSMUST00000088356.4
ENSMUST00000169582.1 |
Gm608
|
predicted gene 608 |
| chr5_+_111417263 | 0.60 |
ENSMUST00000094463.4
|
Mn1
|
meningioma 1 |
| chr11_+_74619594 | 0.60 |
ENSMUST00000100866.2
|
E130309D14Rik
|
RIKEN cDNA E130309D14 gene |
| chr13_+_21754067 | 0.59 |
ENSMUST00000091709.2
|
Hist1h2bn
|
histone cluster 1, H2bn |
| chr15_-_103255433 | 0.59 |
ENSMUST00000075192.6
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr11_+_102393403 | 0.59 |
ENSMUST00000107105.2
ENSMUST00000107102.1 ENSMUST00000107103.1 ENSMUST00000006750.7 |
Rundc3a
|
RUN domain containing 3A |
| chr16_-_20621255 | 0.59 |
ENSMUST00000052939.2
|
Camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr5_-_138264013 | 0.59 |
ENSMUST00000161604.1
ENSMUST00000163268.1 |
BC037034
BC037034
|
cDNA sequence BC037034 cDNA sequence BC037034 |
| chr14_-_69503220 | 0.58 |
ENSMUST00000180059.2
|
Gm21464
|
predicted gene, 21464 |
| chr11_-_116077562 | 0.58 |
ENSMUST00000174822.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr4_-_117872520 | 0.56 |
ENSMUST00000171052.1
ENSMUST00000166325.1 ENSMUST00000106422.2 |
Ccdc24
|
coiled-coil domain containing 24 |
| chr5_+_93206518 | 0.56 |
ENSMUST00000031330.4
|
2010109A12Rik
|
RIKEN cDNA 2010109A12 gene |
| chr11_-_116076986 | 0.55 |
ENSMUST00000153408.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr11_-_74723829 | 0.55 |
ENSMUST00000102520.2
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr7_+_44428938 | 0.55 |
ENSMUST00000127790.1
|
Lrrc4b
|
leucine rich repeat containing 4B |
| chr7_+_99535439 | 0.54 |
ENSMUST00000098266.2
ENSMUST00000179755.1 |
Arrb1
|
arrestin, beta 1 |
| chr11_-_88718165 | 0.53 |
ENSMUST00000107908.1
|
Msi2
|
musashi RNA-binding protein 2 |
| chr11_+_113619318 | 0.53 |
ENSMUST00000146390.2
ENSMUST00000106630.1 |
Sstr2
|
somatostatin receptor 2 |
| chr5_-_138263942 | 0.53 |
ENSMUST00000048421.7
ENSMUST00000164203.1 |
BC037034
BC037034
|
cDNA sequence BC037034 cDNA sequence BC037034 |
| chr6_-_39420281 | 0.52 |
ENSMUST00000114822.1
ENSMUST00000051671.4 |
Mkrn1
|
makorin, ring finger protein, 1 |
| chr3_+_28263205 | 0.52 |
ENSMUST00000159236.2
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr11_+_88718442 | 0.51 |
ENSMUST00000138007.1
|
C030037D09Rik
|
RIKEN cDNA C030037D09 gene |
| chr4_-_84674989 | 0.51 |
ENSMUST00000175800.1
ENSMUST00000176418.1 ENSMUST00000175969.1 ENSMUST00000176370.1 ENSMUST00000176947.1 ENSMUST00000102820.2 ENSMUST00000107198.2 ENSMUST00000175756.1 ENSMUST00000176691.1 |
Bnc2
|
basonuclin 2 |
| chr7_+_126823287 | 0.50 |
ENSMUST00000079423.5
|
Fam57b
|
family with sequence similarity 57, member B |
| chr19_+_55741810 | 0.50 |
ENSMUST00000111657.3
ENSMUST00000061496.9 ENSMUST00000041717.7 ENSMUST00000111662.4 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr3_+_89183131 | 0.50 |
ENSMUST00000140473.1
ENSMUST00000041913.6 |
Fam189b
|
family with sequence similarity 189, member B |
| chr2_+_151842912 | 0.50 |
ENSMUST00000042217.3
|
Rspo4
|
R-spondin family, member 4 |
| chr7_+_57591147 | 0.50 |
ENSMUST00000039697.7
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 |
| chr11_-_69122589 | 0.50 |
ENSMUST00000180487.1
|
9130213A22Rik
|
RIKEN cDNA 9130213A22 gene |
| chr9_-_97018823 | 0.50 |
ENSMUST00000055433.4
|
Spsb4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr7_+_99535652 | 0.49 |
ENSMUST00000032995.8
ENSMUST00000162404.1 |
Arrb1
|
arrestin, beta 1 |
| chr3_+_107896247 | 0.49 |
ENSMUST00000169365.1
|
Gstm5
|
glutathione S-transferase, mu 5 |
| chr1_-_174250976 | 0.48 |
ENSMUST00000061990.4
|
Olfr419
|
olfactory receptor 419 |
| chr3_-_89093358 | 0.48 |
ENSMUST00000090929.5
ENSMUST00000052539.6 |
Rusc1
|
RUN and SH3 domain containing 1 |
| chr14_-_39472825 | 0.48 |
ENSMUST00000168810.2
ENSMUST00000173780.1 ENSMUST00000166968.2 |
Nrg3
|
neuregulin 3 |
| chr7_-_27355944 | 0.47 |
ENSMUST00000003857.6
|
Shkbp1
|
Sh3kbp1 binding protein 1 |
| chr4_+_155962292 | 0.47 |
ENSMUST00000024338.4
|
Fam132a
|
family with sequence similarity 132, member A |
| chr10_-_42583628 | 0.47 |
ENSMUST00000019938.4
|
Nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr2_-_5714490 | 0.46 |
ENSMUST00000044009.7
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr11_-_89302545 | 0.46 |
ENSMUST00000061728.3
|
Nog
|
noggin |
| chr4_-_129491022 | 0.46 |
ENSMUST00000000421.5
|
Tssk3
|
testis-specific serine kinase 3 |
| chr4_+_43632185 | 0.45 |
ENSMUST00000107874.2
|
Npr2
|
natriuretic peptide receptor 2 |
| chr7_-_34654342 | 0.45 |
ENSMUST00000108069.1
|
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chr19_-_6987621 | 0.44 |
ENSMUST00000130048.1
ENSMUST00000025914.6 |
Vegfb
|
vascular endothelial growth factor B |
| chr3_+_103576081 | 0.44 |
ENSMUST00000183637.1
ENSMUST00000117221.2 ENSMUST00000118117.1 ENSMUST00000118563.2 |
Syt6
|
synaptotagmin VI |
| chr11_-_69981242 | 0.44 |
ENSMUST00000108594.1
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr7_+_127746775 | 0.44 |
ENSMUST00000033081.7
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr5_+_136967859 | 0.44 |
ENSMUST00000001790.5
|
Cldn15
|
claudin 15 |
| chr18_-_77565050 | 0.44 |
ENSMUST00000182153.1
ENSMUST00000182146.1 ENSMUST00000026494.7 ENSMUST00000182024.1 |
Rnf165
|
ring finger protein 165 |
| chr6_-_42324640 | 0.44 |
ENSMUST00000031891.8
ENSMUST00000143278.1 |
Fam131b
|
family with sequence similarity 131, member B |
| chrX_-_143933204 | 0.44 |
ENSMUST00000112851.1
ENSMUST00000112856.2 ENSMUST00000033642.3 |
Dcx
|
doublecortin |
| chr1_+_167001457 | 0.43 |
ENSMUST00000126198.1
|
Fam78b
|
family with sequence similarity 78, member B |
| chr11_-_100527896 | 0.43 |
ENSMUST00000107389.1
ENSMUST00000007131.9 |
Acly
|
ATP citrate lyase |
| chr11_+_69981127 | 0.43 |
ENSMUST00000108593.1
|
Ctdnep1
|
CTD nuclear envelope phosphatase 1 |
| chr4_+_127169131 | 0.43 |
ENSMUST00000046659.7
|
Dlgap3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr5_+_93268247 | 0.43 |
ENSMUST00000121127.1
|
Ccng2
|
cyclin G2 |
| chr3_-_88503187 | 0.42 |
ENSMUST00000120377.1
|
Lmna
|
lamin A |
| chr1_+_134289997 | 0.42 |
ENSMUST00000027730.4
|
Myog
|
myogenin |
| chr2_-_17731035 | 0.42 |
ENSMUST00000028080.5
|
Nebl
|
nebulette |
| chr11_+_58640394 | 0.41 |
ENSMUST00000075084.4
|
Trim58
|
tripartite motif-containing 58 |
| chr11_+_96271453 | 0.41 |
ENSMUST00000000010.8
ENSMUST00000174042.1 |
Hoxb9
|
homeobox B9 |
| chr3_-_88503331 | 0.41 |
ENSMUST00000029699.6
|
Lmna
|
lamin A |
| chr2_-_126675461 | 0.41 |
ENSMUST00000103226.3
ENSMUST00000110424.2 |
Gabpb1
|
GA repeat binding protein, beta 1 |
| chr5_-_24540439 | 0.41 |
ENSMUST00000048302.6
ENSMUST00000119657.1 |
Asb10
|
ankyrin repeat and SOCS box-containing 10 |
| chrX_-_7899220 | 0.41 |
ENSMUST00000033497.2
|
Pqbp1
|
polyglutamine binding protein 1 |
| chrX_-_7898950 | 0.41 |
ENSMUST00000115655.1
ENSMUST00000156741.1 |
Pqbp1
|
polyglutamine binding protein 1 |
| chr10_+_80805233 | 0.40 |
ENSMUST00000036016.4
|
Amh
|
anti-Mullerian hormone |
| chr12_+_32378692 | 0.40 |
ENSMUST00000172332.2
|
Ccdc71l
|
coiled-coil domain containing 71 like |
| chr15_-_79932516 | 0.40 |
ENSMUST00000177044.1
ENSMUST00000109615.1 ENSMUST00000089293.4 ENSMUST00000109616.2 |
Cbx7
|
chromobox 7 |
| chr11_-_97699634 | 0.40 |
ENSMUST00000103148.1
ENSMUST00000169807.1 |
Pcgf2
|
polycomb group ring finger 2 |
| chr1_-_180813591 | 0.40 |
ENSMUST00000162118.1
ENSMUST00000159685.1 ENSMUST00000161308.1 |
H3f3a
|
H3 histone, family 3A |
| chr11_+_94990996 | 0.39 |
ENSMUST00000038696.5
|
Ppp1r9b
|
protein phosphatase 1, regulatory subunit 9B |
| chr17_-_27909206 | 0.39 |
ENSMUST00000114848.1
|
Taf11
|
TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr2_-_164171113 | 0.39 |
ENSMUST00000045196.3
|
Kcns1
|
K+ voltage-gated channel, subfamily S, 1 |
| chr7_-_98145472 | 0.39 |
ENSMUST00000098281.2
|
Omp
|
olfactory marker protein |
| chr11_-_98149551 | 0.39 |
ENSMUST00000103143.3
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr4_+_129491190 | 0.39 |
ENSMUST00000106043.2
|
Fam229a
|
family with sequence similarity 229, member A |
| chr1_+_92831614 | 0.39 |
ENSMUST00000045970.6
|
Gpc1
|
glypican 1 |
| chrX_-_7899196 | 0.39 |
ENSMUST00000115654.1
|
Pqbp1
|
polyglutamine binding protein 1 |
| chr17_+_47594629 | 0.38 |
ENSMUST00000182846.1
|
Ccnd3
|
cyclin D3 |
| chr2_-_126675538 | 0.38 |
ENSMUST00000103227.1
ENSMUST00000110425.2 ENSMUST00000089745.4 |
Gabpb1
|
GA repeat binding protein, beta 1 |
| chr7_+_18991245 | 0.37 |
ENSMUST00000130268.1
ENSMUST00000059331.8 ENSMUST00000131087.1 |
Mypop
|
Myb-related transcription factor, partner of profilin |
| chr6_-_31218421 | 0.37 |
ENSMUST00000115107.1
|
AB041803
|
cDNA sequence AB041803 |
| chr11_+_97415527 | 0.37 |
ENSMUST00000121799.1
|
Arhgap23
|
Rho GTPase activating protein 23 |
| chr5_+_64803513 | 0.37 |
ENSMUST00000165536.1
|
Klf3
|
Kruppel-like factor 3 (basic) |
| chr11_+_70657687 | 0.37 |
ENSMUST00000134087.1
ENSMUST00000170716.1 |
Eno3
|
enolase 3, beta muscle |
| chr17_-_87797994 | 0.36 |
ENSMUST00000055221.7
|
Kcnk12
|
potassium channel, subfamily K, member 12 |
| chr14_+_62760496 | 0.36 |
ENSMUST00000181344.1
|
4931440J10Rik
|
RIKEN cDNA 4931440J10 gene |
| chr7_-_101302020 | 0.36 |
ENSMUST00000122116.1
ENSMUST00000120267.1 |
Atg16l2
|
autophagy related 16-like 2 (S. cerevisiae) |
| chr11_-_88718078 | 0.36 |
ENSMUST00000092794.5
|
Msi2
|
musashi RNA-binding protein 2 |
| chr11_-_6606053 | 0.36 |
ENSMUST00000045713.3
|
Nacad
|
NAC alpha domain containing |
| chr6_-_39419757 | 0.36 |
ENSMUST00000146785.1
ENSMUST00000114823.1 |
Mkrn1
|
makorin, ring finger protein, 1 |
| chr4_-_43584386 | 0.36 |
ENSMUST00000107884.2
|
Msmp
|
microseminoprotein, prostate associated |
| chr6_-_42324554 | 0.36 |
ENSMUST00000095974.3
|
Fam131b
|
family with sequence similarity 131, member B |
| chr14_+_54476100 | 0.35 |
ENSMUST00000164766.1
ENSMUST00000164697.1 |
Rem2
|
rad and gem related GTP binding protein 2 |
| chr5_+_137288273 | 0.35 |
ENSMUST00000024099.4
ENSMUST00000085934.3 |
Ache
|
acetylcholinesterase |
| chr5_-_32133045 | 0.35 |
ENSMUST00000031308.6
|
Gm10463
|
predicted gene 10463 |
| chr8_+_85026833 | 0.35 |
ENSMUST00000047281.8
|
2310036O22Rik
|
RIKEN cDNA 2310036O22 gene |
| chr5_-_145201829 | 0.34 |
ENSMUST00000162220.1
ENSMUST00000031632.8 |
Zkscan14
|
zinc finger with KRAB and SCAN domains 14 |
| chr17_+_26941420 | 0.34 |
ENSMUST00000081285.3
ENSMUST00000177932.1 |
Syngap1
|
synaptic Ras GTPase activating protein 1 homolog (rat) |
| chr1_-_180813534 | 0.34 |
ENSMUST00000159789.1
ENSMUST00000081026.4 |
H3f3a
|
H3 histone, family 3A |
| chr6_-_39419967 | 0.34 |
ENSMUST00000122996.1
|
Mkrn1
|
makorin, ring finger protein, 1 |
| chr17_-_35205305 | 0.34 |
ENSMUST00000025266.5
|
Lta
|
lymphotoxin A |
| chr6_-_28134545 | 0.34 |
ENSMUST00000115323.1
|
Grm8
|
glutamate receptor, metabotropic 8 |
| chr1_+_15287259 | 0.34 |
ENSMUST00000175681.1
|
Kcnb2
|
potassium voltage gated channel, Shab-related subfamily, member 2 |
| chr13_+_23746734 | 0.34 |
ENSMUST00000099703.2
|
Hist1h2bb
|
histone cluster 1, H2bb |
| chr2_+_25242929 | 0.34 |
ENSMUST00000114355.1
ENSMUST00000060818.1 |
Rnf208
|
ring finger protein 208 |
| chr17_+_28801090 | 0.34 |
ENSMUST00000004985.9
|
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr17_+_47593444 | 0.33 |
ENSMUST00000182209.1
|
Ccnd3
|
cyclin D3 |
| chr4_+_118428078 | 0.33 |
ENSMUST00000006557.6
ENSMUST00000167636.1 ENSMUST00000102673.4 |
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr4_+_45184815 | 0.33 |
ENSMUST00000134280.1
ENSMUST00000044773.5 |
Frmpd1
|
FERM and PDZ domain containing 1 |
| chr17_+_69969073 | 0.33 |
ENSMUST00000133983.1
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr4_-_133339238 | 0.33 |
ENSMUST00000105906.1
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr5_+_118560719 | 0.33 |
ENSMUST00000100816.4
|
Med13l
|
mediator complex subunit 13-like |
| chr11_-_69920892 | 0.32 |
ENSMUST00000152589.1
ENSMUST00000108612.1 ENSMUST00000108611.1 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr4_+_99656299 | 0.32 |
ENSMUST00000087285.3
|
Foxd3
|
forkhead box D3 |
| chr8_+_121127827 | 0.32 |
ENSMUST00000181609.1
|
Foxl1
|
forkhead box L1 |
| chr19_-_24555819 | 0.31 |
ENSMUST00000112673.2
ENSMUST00000025800.8 |
Pip5k1b
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 beta |
| chr7_-_4778141 | 0.31 |
ENSMUST00000094892.5
|
Il11
|
interleukin 11 |
| chr12_+_111039334 | 0.31 |
ENSMUST00000084968.7
|
Rcor1
|
REST corepressor 1 |
| chr15_+_81235499 | 0.31 |
ENSMUST00000166855.1
|
Mchr1
|
melanin-concentrating hormone receptor 1 |
| chr7_+_29134854 | 0.31 |
ENSMUST00000161522.1
ENSMUST00000159975.1 ENSMUST00000032811.5 ENSMUST00000094617.4 |
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chrX_-_7898864 | 0.31 |
ENSMUST00000154552.1
|
Pqbp1
|
polyglutamine binding protein 1 |
| chr6_-_85502858 | 0.31 |
ENSMUST00000161546.1
ENSMUST00000161078.1 |
Fbxo41
|
F-box protein 41 |
| chr2_-_121271403 | 0.31 |
ENSMUST00000110648.1
|
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr7_-_30534180 | 0.31 |
ENSMUST00000044338.4
|
Arhgap33
|
Rho GTPase activating protein 33 |
| chr6_-_124813065 | 0.31 |
ENSMUST00000149610.2
|
Tpi1
|
triosephosphate isomerase 1 |
| chr10_-_127288851 | 0.30 |
ENSMUST00000156208.1
ENSMUST00000026476.6 |
Mbd6
|
methyl-CpG binding domain protein 6 |
| chr2_-_121271341 | 0.30 |
ENSMUST00000110647.1
|
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr15_+_61987410 | 0.30 |
ENSMUST00000160009.1
|
Myc
|
myelocytomatosis oncogene |
| chr7_-_115824699 | 0.30 |
ENSMUST00000169129.1
|
Sox6
|
SRY-box containing gene 6 |
| chr6_-_142964404 | 0.30 |
ENSMUST00000032421.3
|
St8sia1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr4_-_20778852 | 0.30 |
ENSMUST00000102998.3
|
Nkain3
|
Na+/K+ transporting ATPase interacting 3 |
| chr1_+_82839449 | 0.29 |
ENSMUST00000113444.1
ENSMUST00000063380.4 |
Agfg1
|
ArfGAP with FG repeats 1 |
| chr7_+_19361207 | 0.29 |
ENSMUST00000047621.7
|
Ppp1r13l
|
protein phosphatase 1, regulatory (inhibitor) subunit 13 like |
| chr9_-_98955302 | 0.29 |
ENSMUST00000181706.1
|
Foxl2os
|
forkhead box L2 opposite strand transcript |
| chr5_-_149636164 | 0.29 |
ENSMUST00000076410.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr1_+_180893107 | 0.29 |
ENSMUST00000085797.5
|
Lefty2
|
left-right determination factor 2 |
| chr17_+_47593516 | 0.29 |
ENSMUST00000182874.1
|
Ccnd3
|
cyclin D3 |
| chr15_-_99087817 | 0.29 |
ENSMUST00000064462.3
|
C1ql4
|
complement component 1, q subcomponent-like 4 |
| chr10_-_128645784 | 0.29 |
ENSMUST00000065334.3
|
Ikzf4
|
IKAROS family zinc finger 4 |
| chr4_+_43631935 | 0.29 |
ENSMUST00000030191.8
|
Npr2
|
natriuretic peptide receptor 2 |
| chr10_-_128645965 | 0.29 |
ENSMUST00000133342.1
|
Ikzf4
|
IKAROS family zinc finger 4 |
| chr18_+_46741903 | 0.29 |
ENSMUST00000025357.7
|
Ap3s1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chrX_-_7967817 | 0.29 |
ENSMUST00000033502.7
|
Gata1
|
GATA binding protein 1 |
| chr6_-_113195380 | 0.28 |
ENSMUST00000162280.1
|
Lhfpl4
|
lipoma HMGIC fusion partner-like protein 4 |
| chr7_-_130547358 | 0.28 |
ENSMUST00000160289.1
|
Nsmce4a
|
non-SMC element 4 homolog A (S. cerevisiae) |
| chr9_+_50856924 | 0.28 |
ENSMUST00000174628.1
ENSMUST00000034560.7 ENSMUST00000114437.2 ENSMUST00000175645.1 ENSMUST00000176349.1 ENSMUST00000176798.1 ENSMUST00000175640.1 |
Ppp2r1b
|
protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), beta isoform |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mzf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.3 | 1.0 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.3 | 1.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.3 | 2.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 1.3 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 2.2 | GO:1902033 | regulation of hematopoietic stem cell proliferation(GO:1902033) |
| 0.2 | 1.2 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.2 | 1.2 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.2 | 1.9 | GO:0090343 | positive regulation of cell aging(GO:0090343) |
| 0.2 | 1.5 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.2 | 0.7 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.2 | 2.0 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.2 | 0.3 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.2 | 0.7 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.2 | 1.1 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.2 | 0.5 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.2 | 0.8 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.9 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.4 | GO:0014878 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 | 0.6 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.1 | 0.6 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.5 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.4 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 0.1 | GO:0061324 | canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) |
| 0.1 | 0.4 | GO:0021622 | oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.1 | 0.4 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 1.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.1 | 0.4 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 1.8 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 1.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.6 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.1 | 0.9 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.4 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 1.8 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 0.3 | GO:0061193 | taste bud development(GO:0061193) |
| 0.1 | 0.5 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.3 | GO:0046166 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.6 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.6 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.4 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 0.5 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.4 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.6 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.4 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.1 | 0.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.3 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 1.7 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.2 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.1 | 0.3 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.1 | 0.2 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) neuroblast migration(GO:0097402) |
| 0.1 | 0.7 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.2 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.1 | 0.3 | GO:0046722 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 | 0.3 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 | 0.7 | GO:0021843 | substrate-independent telencephalic tangential migration(GO:0021826) substrate-independent telencephalic tangential interneuron migration(GO:0021843) |
| 0.1 | 0.2 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 | 0.3 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.1 | 0.3 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.1 | GO:0032499 | detection of peptidoglycan(GO:0032499) |
| 0.1 | 0.2 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 0.1 | 0.2 | GO:1901608 | regulation of vesicle transport along microtubule(GO:1901608) |
| 0.1 | 0.2 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.1 | 0.2 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 1.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 1.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.3 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.1 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.2 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 0.2 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 0.1 | 0.2 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.1 | 1.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.6 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.2 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 0.1 | GO:0071608 | macrophage inflammatory protein-1 alpha production(GO:0071608) |
| 0.1 | 0.5 | GO:0030432 | peristalsis(GO:0030432) |
| 0.1 | 0.3 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.1 | 0.3 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.3 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 0.5 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.2 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.1 | 0.2 | GO:0002268 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.4 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 0.3 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.3 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.0 | 0.6 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.2 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.3 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:1904057 | negative regulation of sensory perception of pain(GO:1904057) |
| 0.0 | 0.1 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.3 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.2 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.0 | 0.9 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.1 | GO:0002352 | B cell negative selection(GO:0002352) |
| 0.0 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.4 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.3 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 1.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.0 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.3 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:0070432 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) |
| 0.0 | 0.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.0 | 0.9 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.1 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.2 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.0 | GO:0060264 | regulation of respiratory burst involved in inflammatory response(GO:0060264) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:0035483 | gastric emptying(GO:0035483) |
| 0.0 | 0.1 | GO:0090381 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.6 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.2 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:0070649 | meiotic chromosome movement towards spindle pole(GO:0016344) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 0.1 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.6 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.4 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.0 | 0.1 | GO:1902218 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.3 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.3 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.3 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.5 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.0 | GO:0003169 | coronary vein morphogenesis(GO:0003169) positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.0 | 0.1 | GO:0090005 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 1.3 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.4 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 0.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.0 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 1.2 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.2 | GO:0048617 | foregut morphogenesis(GO:0007440) embryonic foregut morphogenesis(GO:0048617) |
| 0.0 | 0.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.3 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.6 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.0 | 0.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.8 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 1.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.4 | GO:2000726 | negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.0 | 0.2 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.0 | 0.4 | GO:0060384 | innervation(GO:0060384) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.3 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.2 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.1 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.3 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.0 | 0.4 | GO:0009584 | detection of visible light(GO:0009584) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.1 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.3 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.0 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 2.2 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.1 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 0.0 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.0 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.1 | GO:0043084 | penile erection(GO:0043084) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0045060 | negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.2 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.1 | GO:0075522 | IRES-dependent translational initiation(GO:0002192) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.3 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.3 | 2.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 0.9 | GO:0090537 | CERF complex(GO:0090537) |
| 0.2 | 2.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 1.0 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 0.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.7 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 1.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.8 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.2 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 0.7 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 1.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.5 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.8 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 1.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 1.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.4 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 1.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.4 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.2 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.0 | 0.2 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.0 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 2.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.8 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 1.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0005186 | insulin-activated receptor activity(GO:0005009) pheromone activity(GO:0005186) |
| 0.3 | 1.0 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.3 | 1.0 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.3 | 0.9 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 0.3 | 1.2 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.3 | 2.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 0.6 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.2 | 1.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 2.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.2 | 0.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.2 | 0.2 | GO:0031691 | alpha-1A adrenergic receptor binding(GO:0031691) alpha-1B adrenergic receptor binding(GO:0031692) follicle-stimulating hormone receptor binding(GO:0031762) |
| 0.1 | 0.4 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 0.8 | GO:0010851 | cyclase regulator activity(GO:0010851) |
| 0.1 | 1.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.7 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.6 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.5 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 1.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.3 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.1 | 0.5 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 1.8 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.3 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 0.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.8 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.5 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.2 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.1 | 1.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 1.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 1.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 1.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 1.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.3 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.1 | 0.3 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.1 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 1.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.5 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.3 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 2.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 1.1 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.9 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.4 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.0 | 0.1 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 1.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 1.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.2 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 1.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.6 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.0 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 1.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.1 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.5 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 2.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.1 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.8 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 1.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.8 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.1 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.2 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME MAPK TARGETS NUCLEAR EVENTS MEDIATED BY MAP KINASES | Genes involved in MAPK targets/ Nuclear events mediated by MAP kinases |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.2 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.6 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.1 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 2.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |