Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
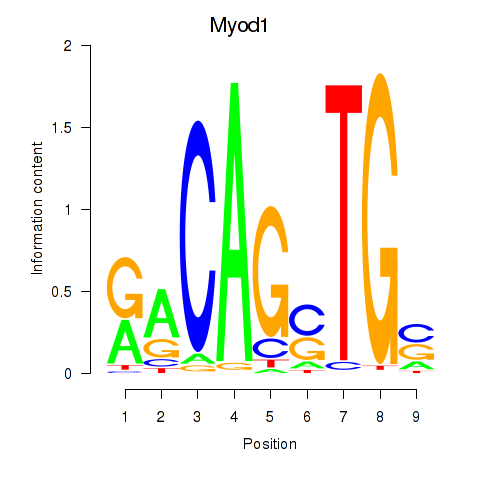
Results for Myod1
Z-value: 2.30
Transcription factors associated with Myod1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Myod1
|
ENSMUSG00000009471.3 | myogenic differentiation 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Myod1 | mm10_v2_chr7_+_46376467_46376480 | 0.67 | 8.7e-06 | Click! |
Activity profile of Myod1 motif
Sorted Z-values of Myod1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_23139064 | 30.79 |
ENSMUST00000033947.8
|
Ank1
|
ankyrin 1, erythroid |
| chr8_+_23139030 | 28.96 |
ENSMUST00000121075.1
|
Ank1
|
ankyrin 1, erythroid |
| chr11_+_58640394 | 16.64 |
ENSMUST00000075084.4
|
Trim58
|
tripartite motif-containing 58 |
| chr12_+_109459843 | 11.48 |
ENSMUST00000173812.1
|
Dlk1
|
delta-like 1 homolog (Drosophila) |
| chr8_+_23139157 | 10.66 |
ENSMUST00000174435.1
|
Ank1
|
ankyrin 1, erythroid |
| chr1_-_132390301 | 10.56 |
ENSMUST00000132435.1
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr2_-_170406501 | 10.48 |
ENSMUST00000154650.1
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr7_+_142441808 | 10.29 |
ENSMUST00000105971.1
ENSMUST00000145287.1 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr12_-_76709997 | 10.19 |
ENSMUST00000166101.1
|
Sptb
|
spectrin beta, erythrocytic |
| chr17_+_36869567 | 10.02 |
ENSMUST00000060524.9
|
Trim10
|
tripartite motif-containing 10 |
| chr11_-_53480178 | 9.25 |
ENSMUST00000104955.2
|
Sowaha
|
sosondowah ankyrin repeat domain family member A |
| chr11_+_115899943 | 9.10 |
ENSMUST00000152171.1
|
Smim5
|
small integral membrane protein 5 |
| chr18_+_34840575 | 9.06 |
ENSMUST00000043484.7
|
Reep2
|
receptor accessory protein 2 |
| chr11_+_115900125 | 8.79 |
ENSMUST00000142089.1
ENSMUST00000131566.1 |
Smim5
|
small integral membrane protein 5 |
| chr1_-_75133866 | 8.71 |
ENSMUST00000027405.4
|
Slc23a3
|
solute carrier family 23 (nucleobase transporters), member 3 |
| chr5_-_107723954 | 8.12 |
ENSMUST00000165344.1
|
Gfi1
|
growth factor independent 1 |
| chr4_+_46039202 | 7.74 |
ENSMUST00000156200.1
|
Tmod1
|
tropomodulin 1 |
| chrX_-_7964166 | 7.64 |
ENSMUST00000128449.1
|
Gata1
|
GATA binding protein 1 |
| chr7_+_127211608 | 7.47 |
ENSMUST00000032910.6
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr17_-_26199008 | 7.43 |
ENSMUST00000142410.1
ENSMUST00000120333.1 ENSMUST00000039113.7 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr7_+_110772604 | 7.43 |
ENSMUST00000005829.6
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr17_+_25471564 | 7.41 |
ENSMUST00000025002.1
|
Tekt4
|
tektin 4 |
| chr9_-_39604124 | 7.27 |
ENSMUST00000042485.4
ENSMUST00000141370.1 |
AW551984
|
expressed sequence AW551984 |
| chr9_+_121777607 | 7.07 |
ENSMUST00000098272.2
|
Klhl40
|
kelch-like 40 |
| chr2_+_153492790 | 7.02 |
ENSMUST00000109783.1
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr3_-_116253467 | 6.89 |
ENSMUST00000090473.5
|
Gpr88
|
G-protein coupled receptor 88 |
| chr11_-_69605829 | 6.81 |
ENSMUST00000047889.6
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr7_+_19411086 | 6.78 |
ENSMUST00000003643.1
|
Ckm
|
creatine kinase, muscle |
| chr6_+_122391379 | 6.54 |
ENSMUST00000043553.3
|
1700063H04Rik
|
RIKEN cDNA 1700063H04 gene |
| chr5_-_73191848 | 6.49 |
ENSMUST00000176910.1
|
Fryl
|
furry homolog-like (Drosophila) |
| chr5_+_123076275 | 6.44 |
ENSMUST00000067505.8
ENSMUST00000111619.3 ENSMUST00000160344.1 |
Tmem120b
|
transmembrane protein 120B |
| chr11_-_54028090 | 6.42 |
ENSMUST00000020586.6
|
Slc22a4
|
solute carrier family 22 (organic cation transporter), member 4 |
| chr16_-_16869255 | 6.39 |
ENSMUST00000075017.4
|
Vpreb1
|
pre-B lymphocyte gene 1 |
| chr16_+_17980565 | 6.36 |
ENSMUST00000075371.3
|
Vpreb2
|
pre-B lymphocyte gene 2 |
| chr7_+_45639964 | 6.24 |
ENSMUST00000148532.1
|
Mamstr
|
MEF2 activating motif and SAP domain containing transcriptional regulator |
| chrX_+_160390684 | 6.09 |
ENSMUST00000112408.2
ENSMUST00000112402.1 ENSMUST00000112401.1 ENSMUST00000112400.1 ENSMUST00000112405.2 ENSMUST00000112404.2 ENSMUST00000146805.1 |
Gpr64
|
G protein-coupled receptor 64 |
| chr9_-_42124276 | 6.00 |
ENSMUST00000060989.8
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing |
| chr2_-_114052804 | 5.80 |
ENSMUST00000090269.6
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr2_-_84822546 | 5.79 |
ENSMUST00000028471.5
|
Smtnl1
|
smoothelin-like 1 |
| chr2_-_164779721 | 5.72 |
ENSMUST00000103095.4
|
Tnnc2
|
troponin C2, fast |
| chr11_+_104577281 | 5.70 |
ENSMUST00000106956.3
|
Myl4
|
myosin, light polypeptide 4 |
| chrX_-_52613936 | 5.68 |
ENSMUST00000114857.1
|
Gpc3
|
glypican 3 |
| chr2_+_103969528 | 5.67 |
ENSMUST00000123437.1
ENSMUST00000163256.1 |
Lmo2
|
LIM domain only 2 |
| chr7_+_142442330 | 5.67 |
ENSMUST00000149529.1
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr3_-_152166230 | 5.62 |
ENSMUST00000046614.9
|
Gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr17_-_35027909 | 5.57 |
ENSMUST00000040151.2
|
Sapcd1
|
suppressor APC domain containing 1 |
| chr11_-_120648104 | 5.50 |
ENSMUST00000026134.2
|
Myadml2
|
myeloid-associated differentiation marker-like 2 |
| chrX_-_52613913 | 5.49 |
ENSMUST00000069360.7
|
Gpc3
|
glypican 3 |
| chr15_-_66831625 | 5.39 |
ENSMUST00000164163.1
|
Sla
|
src-like adaptor |
| chrX_-_142306170 | 5.36 |
ENSMUST00000134825.2
|
Kcne1l
|
potassium voltage-gated channel, Isk-related family, member 1-like, pseudogene |
| chr10_-_80813486 | 5.32 |
ENSMUST00000181039.1
ENSMUST00000180438.1 |
Jsrp1
|
junctional sarcoplasmic reticulum protein 1 |
| chr2_+_91035613 | 5.28 |
ENSMUST00000111445.3
ENSMUST00000111446.3 ENSMUST00000050323.5 |
Rapsn
|
receptor-associated protein of the synapse |
| chr7_+_67952817 | 5.26 |
ENSMUST00000005671.8
|
Igf1r
|
insulin-like growth factor I receptor |
| chr5_-_107726017 | 5.08 |
ENSMUST00000159263.2
|
Gfi1
|
growth factor independent 1 |
| chr11_+_103171081 | 5.05 |
ENSMUST00000042286.5
|
Fmnl1
|
formin-like 1 |
| chr8_-_111691002 | 4.97 |
ENSMUST00000034435.5
|
Ctrb1
|
chymotrypsinogen B1 |
| chr8_-_122460666 | 4.93 |
ENSMUST00000006762.5
|
Snai3
|
snail homolog 3 (Drosophila) |
| chr17_-_48432723 | 4.92 |
ENSMUST00000046549.3
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr16_+_87553313 | 4.79 |
ENSMUST00000026700.7
|
Map3k7cl
|
Map3k7 C-terminal like |
| chr7_-_126447642 | 4.77 |
ENSMUST00000146973.1
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr8_+_94977101 | 4.73 |
ENSMUST00000179619.1
|
Gpr56
|
G protein-coupled receptor 56 |
| chr11_+_104576965 | 4.69 |
ENSMUST00000106957.1
|
Myl4
|
myosin, light polypeptide 4 |
| chrX_+_49470450 | 4.66 |
ENSMUST00000114904.3
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr2_+_122637867 | 4.64 |
ENSMUST00000110512.3
|
AA467197
|
expressed sequence AA467197 |
| chr1_-_96872165 | 4.63 |
ENSMUST00000071985.4
|
Slco4c1
|
solute carrier organic anion transporter family, member 4C1 |
| chr2_+_122637844 | 4.63 |
ENSMUST00000047498.8
|
AA467197
|
expressed sequence AA467197 |
| chrX_-_139871637 | 4.60 |
ENSMUST00000033811.7
ENSMUST00000087401.5 |
Morc4
|
microrchidia 4 |
| chrX_+_49470555 | 4.49 |
ENSMUST00000042444.6
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr2_+_103970115 | 4.48 |
ENSMUST00000111143.1
ENSMUST00000138815.1 |
Lmo2
|
LIM domain only 2 |
| chr10_-_128400448 | 4.48 |
ENSMUST00000167859.1
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr3_-_100489324 | 4.46 |
ENSMUST00000061455.8
|
Fam46c
|
family with sequence similarity 46, member C |
| chr7_-_83735503 | 4.44 |
ENSMUST00000001792.4
|
Il16
|
interleukin 16 |
| chr19_+_47178820 | 4.40 |
ENSMUST00000111808.3
|
Neurl1a
|
neuralized homolog 1A (Drosophila) |
| chrX_+_101449078 | 4.34 |
ENSMUST00000033674.5
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr19_+_60755947 | 4.33 |
ENSMUST00000088237.4
|
Nanos1
|
nanos homolog 1 (Drosophila) |
| chr18_+_60963517 | 4.33 |
ENSMUST00000115295.2
ENSMUST00000039904.6 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr9_+_107975529 | 4.30 |
ENSMUST00000035216.4
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr5_+_105415738 | 4.28 |
ENSMUST00000112707.1
|
Lrrc8b
|
leucine rich repeat containing 8 family, member B |
| chr11_-_107716517 | 4.23 |
ENSMUST00000021065.5
|
Cacng1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr2_-_27072175 | 4.19 |
ENSMUST00000009358.2
|
Tmem8c
|
transmembrane protein 8C |
| chr4_-_43523388 | 4.15 |
ENSMUST00000107913.3
ENSMUST00000030184.5 |
Tpm2
|
tropomyosin 2, beta |
| chr4_-_63172118 | 4.11 |
ENSMUST00000030042.2
|
Kif12
|
kinesin family member 12 |
| chr1_+_135132693 | 4.05 |
ENSMUST00000049449.4
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr2_+_103970221 | 3.94 |
ENSMUST00000111140.2
ENSMUST00000111139.2 |
Lmo2
|
LIM domain only 2 |
| chr4_+_45184815 | 3.93 |
ENSMUST00000134280.1
ENSMUST00000044773.5 |
Frmpd1
|
FERM and PDZ domain containing 1 |
| chr2_+_152911311 | 3.91 |
ENSMUST00000028970.7
|
Mylk2
|
myosin, light polypeptide kinase 2, skeletal muscle |
| chr9_-_57836706 | 3.88 |
ENSMUST00000164010.1
ENSMUST00000171444.1 ENSMUST00000098686.3 |
Arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr13_-_55528511 | 3.81 |
ENSMUST00000047877.4
|
Dok3
|
docking protein 3 |
| chr1_+_129273344 | 3.81 |
ENSMUST00000073527.6
ENSMUST00000040311.7 |
Thsd7b
|
thrombospondin, type I, domain containing 7B |
| chr4_-_133872997 | 3.79 |
ENSMUST00000137486.2
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr9_-_21963568 | 3.76 |
ENSMUST00000006397.5
|
Epor
|
erythropoietin receptor |
| chr7_-_25005895 | 3.72 |
ENSMUST00000102858.3
ENSMUST00000080882.6 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr1_+_75507077 | 3.65 |
ENSMUST00000037330.4
|
Inha
|
inhibin alpha |
| chr4_-_43523746 | 3.60 |
ENSMUST00000150592.1
|
Tpm2
|
tropomyosin 2, beta |
| chr1_-_75506331 | 3.57 |
ENSMUST00000113567.2
ENSMUST00000113565.2 |
Obsl1
|
obscurin-like 1 |
| chr9_+_30942541 | 3.57 |
ENSMUST00000068135.6
|
Adamts8
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 8 |
| chr12_-_115790884 | 3.53 |
ENSMUST00000081809.5
|
Ighv1-73
|
immunoglobulin heavy variable 1-73 |
| chr10_+_14523062 | 3.51 |
ENSMUST00000096020.5
|
Gm10335
|
predicted gene 10335 |
| chr1_+_130731963 | 3.50 |
ENSMUST00000039323.6
|
AA986860
|
expressed sequence AA986860 |
| chr4_+_43957401 | 3.50 |
ENSMUST00000030202.7
|
Glipr2
|
GLI pathogenesis-related 2 |
| chrX_+_48519245 | 3.45 |
ENSMUST00000033430.2
|
Rab33a
|
RAB33A, member of RAS oncogene family |
| chr17_+_47505043 | 3.44 |
ENSMUST00000182129.1
ENSMUST00000171031.1 |
Ccnd3
|
cyclin D3 |
| chr13_-_117025505 | 3.43 |
ENSMUST00000022239.6
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr1_+_135836380 | 3.42 |
ENSMUST00000178204.1
|
Tnnt2
|
troponin T2, cardiac |
| chr17_+_47505117 | 3.38 |
ENSMUST00000183044.1
ENSMUST00000037333.10 |
Ccnd3
|
cyclin D3 |
| chr17_+_47505149 | 3.33 |
ENSMUST00000183177.1
ENSMUST00000182848.1 |
Ccnd3
|
cyclin D3 |
| chr13_+_46418266 | 3.31 |
ENSMUST00000037923.3
|
Rbm24
|
RNA binding motif protein 24 |
| chr7_-_19421326 | 3.31 |
ENSMUST00000047020.1
|
A930016O22Rik
|
RIKEN cDNA A930016O22 gene |
| chr7_+_45216671 | 3.29 |
ENSMUST00000134420.1
|
Tead2
|
TEA domain family member 2 |
| chr1_+_164062070 | 3.20 |
ENSMUST00000097491.3
ENSMUST00000027871.7 |
Sell
|
selectin, lymphocyte |
| chr7_-_142969238 | 3.19 |
ENSMUST00000009392.4
ENSMUST00000121862.1 |
Ascl2
|
achaete-scute complex homolog 2 (Drosophila) |
| chr11_+_32205483 | 3.17 |
ENSMUST00000121182.1
|
Snrnp25
|
small nuclear ribonucleoprotein 25 (U11/U12) |
| chr17_-_27167759 | 3.16 |
ENSMUST00000025046.2
|
Ip6k3
|
inositol hexaphosphate kinase 3 |
| chr3_+_103074009 | 3.15 |
ENSMUST00000090715.6
|
Ampd1
|
adenosine monophosphate deaminase 1 |
| chr17_+_47505211 | 3.13 |
ENSMUST00000182935.1
ENSMUST00000182506.1 |
Ccnd3
|
cyclin D3 |
| chr4_-_133967235 | 3.09 |
ENSMUST00000123234.1
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr3_+_95588990 | 3.06 |
ENSMUST00000177399.1
|
Golph3l
|
golgi phosphoprotein 3-like |
| chr12_+_32378692 | 3.03 |
ENSMUST00000172332.2
|
Ccdc71l
|
coiled-coil domain containing 71 like |
| chr11_+_32205411 | 3.02 |
ENSMUST00000039601.3
ENSMUST00000149043.1 |
Snrnp25
|
small nuclear ribonucleoprotein 25 (U11/U12) |
| chr6_+_30639218 | 2.97 |
ENSMUST00000031806.9
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr15_-_78773452 | 2.94 |
ENSMUST00000018313.5
|
Mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr7_-_140900401 | 2.93 |
ENSMUST00000026561.8
|
Cox8b
|
cytochrome c oxidase subunit VIIIb |
| chr4_-_140845770 | 2.88 |
ENSMUST00000026378.3
|
Padi1
|
peptidyl arginine deiminase, type I |
| chr13_+_91461050 | 2.88 |
ENSMUST00000004094.8
ENSMUST00000042122.8 |
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr7_-_44524642 | 2.88 |
ENSMUST00000165208.2
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr8_+_45885479 | 2.87 |
ENSMUST00000034053.5
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr8_-_105471481 | 2.84 |
ENSMUST00000014990.6
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr11_-_5803733 | 2.84 |
ENSMUST00000020768.3
|
Pgam2
|
phosphoglycerate mutase 2 |
| chr7_+_142471838 | 2.83 |
ENSMUST00000038946.2
|
Lsp1
|
lymphocyte specific 1 |
| chr15_-_77022632 | 2.81 |
ENSMUST00000019037.8
ENSMUST00000169226.1 |
Mb
|
myoglobin |
| chr14_-_62292959 | 2.80 |
ENSMUST00000063169.8
|
Dleu7
|
deleted in lymphocytic leukemia, 7 |
| chr5_+_91517615 | 2.77 |
ENSMUST00000040576.9
|
Parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chrX_-_53114530 | 2.76 |
ENSMUST00000114843.2
|
Plac1
|
placental specific protein 1 |
| chr11_+_69965396 | 2.75 |
ENSMUST00000018713.6
|
Cldn7
|
claudin 7 |
| chr6_-_125494754 | 2.75 |
ENSMUST00000032492.8
|
Cd9
|
CD9 antigen |
| chr5_-_67847400 | 2.74 |
ENSMUST00000113652.1
ENSMUST00000113651.1 ENSMUST00000037380.8 |
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
| chr2_-_33431324 | 2.72 |
ENSMUST00000113158.1
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chr4_+_43957678 | 2.72 |
ENSMUST00000107855.1
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr11_-_79146407 | 2.71 |
ENSMUST00000018478.4
ENSMUST00000108264.1 |
Ksr1
|
kinase suppressor of ras 1 |
| chr8_-_40634750 | 2.66 |
ENSMUST00000173957.1
|
Mtmr7
|
myotubularin related protein 7 |
| chr7_+_142472080 | 2.65 |
ENSMUST00000105966.1
|
Lsp1
|
lymphocyte specific 1 |
| chr11_+_96929367 | 2.64 |
ENSMUST00000062172.5
|
Prr15l
|
proline rich 15-like |
| chr3_-_126998408 | 2.63 |
ENSMUST00000182764.1
ENSMUST00000044443.8 |
Ank2
|
ankyrin 2, brain |
| chr4_-_43523595 | 2.63 |
ENSMUST00000107914.3
|
Tpm2
|
tropomyosin 2, beta |
| chr4_-_135573623 | 2.62 |
ENSMUST00000105855.1
|
Grhl3
|
grainyhead-like 3 (Drosophila) |
| chr4_-_137430517 | 2.55 |
ENSMUST00000102522.4
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr5_+_34989473 | 2.54 |
ENSMUST00000114284.1
ENSMUST00000114285.1 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr3_+_146404631 | 2.51 |
ENSMUST00000106153.2
ENSMUST00000039021.4 ENSMUST00000106151.1 ENSMUST00000149262.1 |
Ssx2ip
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr11_-_120551126 | 2.50 |
ENSMUST00000026121.2
|
Ppp1r27
|
protein phosphatase 1, regulatory subunit 27 |
| chr5_-_67847360 | 2.50 |
ENSMUST00000072971.6
|
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
| chr13_+_23544052 | 2.50 |
ENSMUST00000075558.2
|
Hist1h3f
|
histone cluster 1, H3f |
| chr3_+_137864487 | 2.50 |
ENSMUST00000041045.7
|
H2afz
|
H2A histone family, member Z |
| chr11_+_87760533 | 2.49 |
ENSMUST00000039627.5
ENSMUST00000100644.3 |
Bzrap1
|
benzodiazepine receptor associated protein 1 |
| chr4_+_115088708 | 2.49 |
ENSMUST00000171877.1
ENSMUST00000177647.1 ENSMUST00000106548.2 ENSMUST00000030488.2 |
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr5_+_115011111 | 2.48 |
ENSMUST00000031530.5
|
Sppl3
|
signal peptide peptidase 3 |
| chr7_+_30422389 | 2.47 |
ENSMUST00000108175.1
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr1_+_86021935 | 2.46 |
ENSMUST00000052854.6
ENSMUST00000125083.1 ENSMUST00000152501.1 ENSMUST00000113344.1 ENSMUST00000130504.1 ENSMUST00000153247.2 |
Spata3
|
spermatogenesis associated 3 |
| chr1_+_180935022 | 2.46 |
ENSMUST00000037361.8
|
Lefty1
|
left right determination factor 1 |
| chr11_-_70322520 | 2.45 |
ENSMUST00000019051.2
|
Alox12e
|
arachidonate lipoxygenase, epidermal |
| chr7_-_131410325 | 2.45 |
ENSMUST00000154602.1
|
Ikzf5
|
IKAROS family zinc finger 5 |
| chr5_-_24351604 | 2.45 |
ENSMUST00000036092.7
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr2_+_174760619 | 2.43 |
ENSMUST00000029030.2
|
Edn3
|
endothelin 3 |
| chr7_+_122289297 | 2.40 |
ENSMUST00000064989.5
ENSMUST00000064921.4 |
Prkcb
|
protein kinase C, beta |
| chr4_-_133967296 | 2.37 |
ENSMUST00000105893.1
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr15_+_78926720 | 2.36 |
ENSMUST00000089377.5
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr9_-_112187766 | 2.36 |
ENSMUST00000111872.2
ENSMUST00000164754.2 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr3_+_95588928 | 2.35 |
ENSMUST00000177390.1
ENSMUST00000098861.4 ENSMUST00000060323.5 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr10_-_29699379 | 2.34 |
ENSMUST00000092620.4
|
Gm10275
|
predicted pseudogene 10275 |
| chrX_-_36989656 | 2.33 |
ENSMUST00000060474.7
ENSMUST00000053456.4 ENSMUST00000115239.3 |
Sept6
|
septin 6 |
| chr16_+_48994185 | 2.32 |
ENSMUST00000117994.1
ENSMUST00000048374.5 |
C330027C09Rik
|
RIKEN cDNA C330027C09 gene |
| chr2_-_62483637 | 2.31 |
ENSMUST00000136686.1
ENSMUST00000102733.3 |
Gcg
|
glucagon |
| chr2_+_156840966 | 2.31 |
ENSMUST00000109564.1
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr14_-_20496780 | 2.31 |
ENSMUST00000022353.3
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr3_+_146404978 | 2.30 |
ENSMUST00000129978.1
|
Ssx2ip
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr7_-_127993831 | 2.29 |
ENSMUST00000033056.3
|
Pycard
|
PYD and CARD domain containing |
| chr9_-_121792478 | 2.24 |
ENSMUST00000035110.4
|
Hhatl
|
hedgehog acyltransferase-like |
| chrX_+_159708593 | 2.21 |
ENSMUST00000080394.6
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr3_+_95588960 | 2.20 |
ENSMUST00000176674.1
ENSMUST00000177389.1 ENSMUST00000176755.1 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr2_-_153529941 | 2.20 |
ENSMUST00000035346.7
|
8430427H17Rik
|
RIKEN cDNA 8430427H17 gene |
| chr17_-_35700520 | 2.19 |
ENSMUST00000119825.1
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr11_+_101316917 | 2.17 |
ENSMUST00000151385.1
|
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr1_+_78310295 | 2.17 |
ENSMUST00000036172.8
|
Sgpp2
|
sphingosine-1-phosphate phosphotase 2 |
| chr3_-_108210438 | 2.16 |
ENSMUST00000117784.1
ENSMUST00000119650.1 ENSMUST00000117409.1 |
Atxn7l2
|
ataxin 7-like 2 |
| chr4_+_98923908 | 2.14 |
ENSMUST00000169053.1
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr7_-_98145472 | 2.12 |
ENSMUST00000098281.2
|
Omp
|
olfactory marker protein |
| chr5_-_69542622 | 2.12 |
ENSMUST00000031045.6
|
Yipf7
|
Yip1 domain family, member 7 |
| chr1_+_87205799 | 2.11 |
ENSMUST00000027470.7
|
Chrng
|
cholinergic receptor, nicotinic, gamma polypeptide |
| chr10_+_128909866 | 2.10 |
ENSMUST00000026407.7
|
Cd63
|
CD63 antigen |
| chr5_-_135251209 | 2.10 |
ENSMUST00000062572.2
|
Fzd9
|
frizzled homolog 9 (Drosophila) |
| chr17_-_23684019 | 2.09 |
ENSMUST00000085989.5
|
Cldn9
|
claudin 9 |
| chr11_+_101316200 | 2.05 |
ENSMUST00000142640.1
ENSMUST00000019470.7 |
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr11_+_3983636 | 2.03 |
ENSMUST00000078757.1
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chrX_+_140956892 | 2.03 |
ENSMUST00000112971.1
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr6_-_29212240 | 2.01 |
ENSMUST00000160878.1
ENSMUST00000078155.5 |
Impdh1
|
inosine 5'-phosphate dehydrogenase 1 |
| chr13_-_97747373 | 2.01 |
ENSMUST00000123535.1
|
5330416C01Rik
|
RIKEN cDNA 5330416C01 gene |
| chr1_-_87156127 | 2.00 |
ENSMUST00000160810.1
|
Ecel1
|
endothelin converting enzyme-like 1 |
| chr3_+_146404844 | 1.96 |
ENSMUST00000106149.1
|
Ssx2ip
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr5_-_137212389 | 1.93 |
ENSMUST00000179412.1
|
A630081J09Rik
|
RIKEN cDNA A630081J09 gene |
| chr11_+_96929260 | 1.92 |
ENSMUST00000054311.5
ENSMUST00000107636.3 |
Prr15l
|
proline rich 15-like |
| chr3_+_88616133 | 1.92 |
ENSMUST00000176500.1
ENSMUST00000177498.1 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Myod1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 13.2 | GO:0070103 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 2.5 | 7.6 | GO:0030221 | basophil differentiation(GO:0030221) |
| 2.2 | 11.2 | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 2.2 | 8.7 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 1.9 | 81.5 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 1.7 | 6.9 | GO:0061743 | motor learning(GO:0061743) |
| 1.6 | 9.4 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 1.5 | 9.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 1.5 | 6.0 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 1.5 | 6.0 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) positive regulation of endocytic recycling(GO:2001137) |
| 1.5 | 4.5 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 1.5 | 7.4 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 1.4 | 7.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 1.3 | 12.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 1.3 | 5.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 1.3 | 3.9 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 1.2 | 3.6 | GO:0071846 | actin filament debranching(GO:0071846) |
| 1.2 | 3.6 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 1.1 | 3.4 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 1.0 | 12.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 1.0 | 6.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.9 | 5.5 | GO:0014826 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.9 | 1.8 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.8 | 5.0 | GO:0030240 | skeletal myofibril assembly(GO:0014866) skeletal muscle thin filament assembly(GO:0030240) |
| 0.8 | 16.8 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.7 | 3.6 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.7 | 1.5 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.7 | 0.7 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.7 | 2.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.7 | 2.7 | GO:0060032 | notochord regression(GO:0060032) |
| 0.7 | 5.2 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.6 | 6.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.6 | 2.6 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.6 | 6.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.6 | 29.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.6 | 6.4 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.6 | 8.3 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.6 | 1.9 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.6 | 3.7 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.6 | 2.4 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.6 | 1.8 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.6 | 5.4 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.6 | 3.5 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.6 | 2.9 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.6 | 2.8 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.5 | 2.7 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.5 | 1.6 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.5 | 3.2 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.5 | 5.7 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.5 | 1.5 | GO:2000978 | auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.5 | 1.5 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.5 | 2.5 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.5 | 5.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.5 | 1.5 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.5 | 3.8 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.5 | 1.4 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.5 | 2.7 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.5 | 3.6 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.4 | 2.2 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 0.4 | 5.3 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.4 | 4.4 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.4 | 3.9 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.4 | 2.1 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.4 | 2.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.4 | 2.8 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.4 | 1.2 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.4 | 2.8 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.4 | 2.4 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.4 | 10.5 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.4 | 7.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.4 | 4.3 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.4 | 1.8 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.4 | 4.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.3 | 1.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.3 | 1.3 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.3 | 2.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.3 | 1.0 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.3 | 3.8 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.3 | 2.5 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.3 | 1.2 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.3 | 5.9 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.3 | 0.6 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.3 | 2.6 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.3 | 1.4 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.3 | 0.8 | GO:0072061 | chemoattraction of axon(GO:0061642) inner medullary collecting duct development(GO:0072061) |
| 0.3 | 1.6 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.3 | 3.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.2 | 1.0 | GO:0042125 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.2 | 4.4 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.2 | 2.5 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.2 | 0.7 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.2 | 1.9 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.2 | 0.7 | GO:1901675 | response to methylglyoxal(GO:0051595) negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.2 | 1.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) positive regulation of saliva secretion(GO:0046878) |
| 0.2 | 2.9 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.2 | 0.7 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.2 | 1.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.2 | 2.2 | GO:0042637 | catagen(GO:0042637) |
| 0.2 | 0.9 | GO:0009816 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.2 | 0.9 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.2 | 1.0 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.2 | 2.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 4.3 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.2 | 4.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.2 | 1.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 5.3 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.2 | 7.4 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.2 | 5.2 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.2 | 0.7 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.2 | 1.9 | GO:0046959 | habituation(GO:0046959) |
| 0.2 | 3.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.2 | 3.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.2 | 1.8 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.2 | 0.9 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.2 | 0.3 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.2 | 4.2 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.2 | 9.3 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.2 | 0.3 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.2 | 2.8 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 1.5 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 5.8 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.1 | 1.2 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.1 | 2.2 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.1 | 0.4 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.1 | 2.1 | GO:0051195 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.1 | 1.8 | GO:0060159 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 1.8 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.1 | 1.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 5.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.8 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 1.9 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 12.8 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.1 | 2.5 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 5.2 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 4.5 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.1 | 0.7 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 3.2 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.7 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.8 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.1 | 0.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.6 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 1.0 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.2 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 0.8 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 3.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.4 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.1 | GO:0099548 | trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 0.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 3.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.6 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 2.1 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.1 | 2.2 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 1.7 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.1 | 1.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.1 | 1.2 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.4 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 1.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 1.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.5 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.4 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.5 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.1 | GO:1901382 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.1 | 2.0 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.1 | 2.4 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 0.6 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.9 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 0.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.9 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 | 1.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 2.3 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.1 | 0.1 | GO:1903998 | regulation of eating behavior(GO:1903998) negative regulation of eating behavior(GO:1903999) |
| 0.1 | 2.8 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 1.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 9.6 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 0.3 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 3.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.5 | GO:0042168 | heme metabolic process(GO:0042168) |
| 0.0 | 1.3 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.4 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.5 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.3 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.2 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.6 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 2.3 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.0 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.9 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.8 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.7 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 1.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.2 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.1 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.0 | 1.8 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 | 2.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.4 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.3 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 1.7 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 2.1 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.3 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.7 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.9 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.0 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 79.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 1.4 | 25.1 | GO:0005861 | troponin complex(GO:0005861) |
| 1.2 | 3.6 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 1.1 | 3.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 1.1 | 4.3 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 1.1 | 9.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.1 | 4.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.8 | 4.8 | GO:0031673 | H zone(GO:0031673) |
| 0.8 | 10.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.8 | 2.3 | GO:0005940 | septin ring(GO:0005940) |
| 0.6 | 2.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.6 | 3.6 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.6 | 5.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.6 | 2.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.6 | 11.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.5 | 6.5 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.5 | 2.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.5 | 29.6 | GO:0031672 | A band(GO:0031672) |
| 0.4 | 2.5 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.4 | 6.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.4 | 2.5 | GO:0001740 | Barr body(GO:0001740) |
| 0.3 | 2.7 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.3 | 1.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.3 | 2.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 2.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.3 | 1.8 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 0.8 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 7.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.2 | 0.7 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.2 | 0.7 | GO:0034774 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.2 | 13.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 7.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 6.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 3.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 2.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 1.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 21.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 0.7 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.2 | 8.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 8.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.7 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 1.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 3.4 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.6 | GO:0070820 | cytolytic granule(GO:0044194) tertiary granule(GO:0070820) |
| 0.1 | 1.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 5.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 7.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 3.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 11.5 | GO:0031674 | I band(GO:0031674) |
| 0.1 | 0.9 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 1.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 6.1 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 1.8 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 1.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 3.8 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 0.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 0.9 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 19.2 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 2.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 3.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 13.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 1.4 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.9 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 1.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 5.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 4.8 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 4.0 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.4 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 2.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 1.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 70.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.6 | 16.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 1.4 | 5.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.3 | 12.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.2 | 18.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.1 | 6.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.1 | 6.4 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.9 | 2.8 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.9 | 9.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.9 | 4.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.9 | 8.5 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.8 | 4.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.8 | 2.4 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.7 | 2.8 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.7 | 7.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.7 | 5.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.6 | 3.2 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.6 | 1.9 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.6 | 2.5 | GO:0038100 | nodal binding(GO:0038100) |
| 0.6 | 2.5 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.6 | 2.9 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.6 | 3.9 | GO:0032027 | myosin light chain kinase activity(GO:0004687) myosin light chain binding(GO:0032027) |
| 0.5 | 6.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.5 | 2.7 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.5 | 5.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.5 | 2.0 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.5 | 27.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.5 | 2.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.5 | 7.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.5 | 4.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.5 | 4.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.4 | 1.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.4 | 3.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.4 | 4.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.4 | 4.8 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.4 | 11.0 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.4 | 9.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.4 | 1.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.4 | 2.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 11.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 4.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 5.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 2.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.3 | 4.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.3 | 3.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 5.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 1.2 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.3 | 0.9 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.3 | 1.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.3 | 1.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.3 | 1.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.3 | 1.1 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.3 | 1.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 4.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 1.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 2.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.2 | 8.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 12.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 12.9 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 1.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 0.7 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.2 | 9.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.5 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 1.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.2 | 0.8 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.2 | 1.0 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.2 | 1.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 1.7 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 1.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 3.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 3.2 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.2 | 1.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 1.0 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.2 | 1.0 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 1.6 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.2 | 2.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 3.3 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 1.0 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 3.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.9 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 5.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 4.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 3.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.0 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 1.2 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 1.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 4.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 5.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 2.1 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 3.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.3 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 3.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 3.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 4.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 4.4 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 24.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 3.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 3.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 3.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 2.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 1.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 4.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 6.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.7 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.5 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 1.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 3.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 2.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 4.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 1.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 3.2 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 2.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 2.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 3.3 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.9 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 1.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.3 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 3.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.7 | GO:0017022 | myosin binding(GO:0017022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 19.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 14.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 3.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 5.8 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.2 | 4.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.2 | 5.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 6.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 12.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 6.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 0.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 4.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 5.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 5.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.7 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 2.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 5.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 1.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 6.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.1 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 1.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.9 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 3.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 1.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 5.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 3.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 5.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 1.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 79.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.1 | 56.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.6 | 12.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.5 | 11.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.5 | 6.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.4 | 4.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.4 | 3.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.4 | 4.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.3 | 17.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.3 | 13.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.3 | 10.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 5.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 4.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 8.3 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 2.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 4.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 6.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 4.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 4.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 2.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 1.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.1 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 1.6 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 2.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 2.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 8.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 4.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 3.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.0 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.7 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 1.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 4.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 5.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |