Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
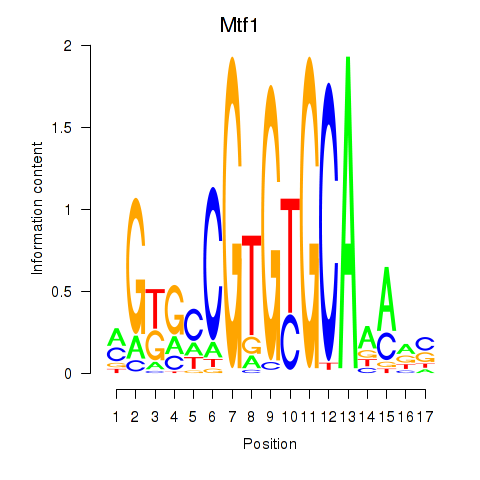
Results for Mtf1
Z-value: 1.55
Transcription factors associated with Mtf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mtf1
|
ENSMUSG00000028890.7 | metal response element binding transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mtf1 | mm10_v2_chr4_+_124802543_124802678 | 0.53 | 9.7e-04 | Click! |
Activity profile of Mtf1 motif
Sorted Z-values of Mtf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_94172618 | 25.30 |
ENSMUST00000034214.6
|
Mt2
|
metallothionein 2 |
| chr8_+_94179089 | 17.13 |
ENSMUST00000034215.6
|
Mt1
|
metallothionein 1 |
| chr9_+_98490522 | 13.23 |
ENSMUST00000035029.2
|
Rbp2
|
retinol binding protein 2, cellular |
| chr6_+_90619241 | 4.59 |
ENSMUST00000032177.8
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr9_-_14381242 | 4.40 |
ENSMUST00000167549.1
|
Endod1
|
endonuclease domain containing 1 |
| chr1_-_169531343 | 4.37 |
ENSMUST00000028000.7
|
Nuf2
|
NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr9_-_56635624 | 3.95 |
ENSMUST00000114256.1
|
Lingo1
|
leucine rich repeat and Ig domain containing 1 |
| chr1_+_75400070 | 3.89 |
ENSMUST00000113589.1
|
Speg
|
SPEG complex locus |
| chr1_-_169531447 | 3.72 |
ENSMUST00000111368.1
|
Nuf2
|
NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr4_+_103619580 | 3.32 |
ENSMUST00000106827.1
|
Dab1
|
disabled 1 |
| chr3_+_107895916 | 3.25 |
ENSMUST00000172247.1
ENSMUST00000167387.1 |
Gstm5
|
glutathione S-transferase, mu 5 |
| chr3_+_107895821 | 3.09 |
ENSMUST00000004134.4
|
Gstm5
|
glutathione S-transferase, mu 5 |
| chr10_+_20952547 | 3.03 |
ENSMUST00000105525.4
|
Ahi1
|
Abelson helper integration site 1 |
| chr3_+_107896247 | 2.88 |
ENSMUST00000169365.1
|
Gstm5
|
glutathione S-transferase, mu 5 |
| chr1_-_167393826 | 2.86 |
ENSMUST00000028005.2
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr5_-_34637107 | 2.76 |
ENSMUST00000124668.1
ENSMUST00000001109.4 ENSMUST00000155577.1 ENSMUST00000114329.1 |
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr19_-_4615453 | 2.76 |
ENSMUST00000053597.2
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr2_+_119112793 | 2.69 |
ENSMUST00000140939.1
ENSMUST00000028795.3 |
Rad51
|
RAD51 homolog |
| chr5_-_34637203 | 2.67 |
ENSMUST00000114331.3
|
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr7_+_43440782 | 2.66 |
ENSMUST00000040227.1
|
Cldnd2
|
claudin domain containing 2 |
| chr19_-_4615647 | 2.60 |
ENSMUST00000113822.2
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr11_+_63133068 | 2.52 |
ENSMUST00000108700.1
|
Pmp22
|
peripheral myelin protein 22 |
| chr4_-_43040279 | 2.43 |
ENSMUST00000107958.1
ENSMUST00000107959.1 ENSMUST00000152846.1 |
Fam214b
|
family with sequence similarity 214, member B |
| chr1_+_135729147 | 2.30 |
ENSMUST00000027677.7
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr11_+_63132569 | 2.27 |
ENSMUST00000108701.1
|
Pmp22
|
peripheral myelin protein 22 |
| chr4_-_126163470 | 2.26 |
ENSMUST00000097891.3
|
Sh3d21
|
SH3 domain containing 21 |
| chr4_+_129985098 | 2.15 |
ENSMUST00000106017.1
ENSMUST00000121049.1 |
Bai2
|
brain-specific angiogenesis inhibitor 2 |
| chr11_-_32222233 | 1.91 |
ENSMUST00000150381.1
ENSMUST00000144902.1 ENSMUST00000020524.8 |
Rhbdf1
|
rhomboid family 1 (Drosophila) |
| chr2_+_29889720 | 1.91 |
ENSMUST00000113767.1
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr3_-_89960562 | 1.89 |
ENSMUST00000069805.7
|
Atp8b2
|
ATPase, class I, type 8B, member 2 |
| chr9_-_65391652 | 1.81 |
ENSMUST00000068307.3
|
Kbtbd13
|
kelch repeat and BTB (POZ) domain containing 13 |
| chr5_+_105415738 | 1.79 |
ENSMUST00000112707.1
|
Lrrc8b
|
leucine rich repeat containing 8 family, member B |
| chr2_+_29889785 | 1.64 |
ENSMUST00000113763.1
ENSMUST00000113757.1 ENSMUST00000113756.1 ENSMUST00000133233.1 ENSMUST00000113759.2 ENSMUST00000113755.1 ENSMUST00000137558.1 ENSMUST00000046571.7 |
Odf2
|
outer dense fiber of sperm tails 2 |
| chr19_-_4201591 | 1.55 |
ENSMUST00000025740.6
|
Rad9a
|
RAD9 homolog A |
| chr3_-_121532271 | 1.48 |
ENSMUST00000039197.7
|
Slc44a3
|
solute carrier family 44, member 3 |
| chr4_-_93335510 | 1.42 |
ENSMUST00000066774.4
|
Tusc1
|
tumor suppressor candidate 1 |
| chr8_+_70905970 | 1.41 |
ENSMUST00000019405.2
|
Map1s
|
microtubule-associated protein 1S |
| chrX_+_6873484 | 1.40 |
ENSMUST00000145302.1
|
Dgkk
|
diacylglycerol kinase kappa |
| chr4_+_129984833 | 1.29 |
ENSMUST00000120204.1
|
Bai2
|
brain-specific angiogenesis inhibitor 2 |
| chr7_+_119794102 | 1.26 |
ENSMUST00000084644.2
|
2610020H08Rik
|
RIKEN cDNA 2610020H08 gene |
| chr5_-_99978914 | 1.25 |
ENSMUST00000112939.3
ENSMUST00000171786.1 ENSMUST00000072750.6 ENSMUST00000019128.8 ENSMUST00000172361.1 |
Hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr6_+_18848571 | 1.24 |
ENSMUST00000056398.8
|
Naa38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr4_-_126162619 | 1.21 |
ENSMUST00000094760.2
|
Sh3d21
|
SH3 domain containing 21 |
| chr1_-_37719782 | 1.20 |
ENSMUST00000160589.1
|
2010300C02Rik
|
RIKEN cDNA 2010300C02 gene |
| chr7_+_119793987 | 1.15 |
ENSMUST00000033218.8
ENSMUST00000106520.1 |
2610020H08Rik
|
RIKEN cDNA 2610020H08 gene |
| chr7_-_109865586 | 1.13 |
ENSMUST00000007423.5
ENSMUST00000106728.2 ENSMUST00000106729.1 |
Scube2
|
signal peptide, CUB domain, EGF-like 2 |
| chr4_-_152152207 | 1.13 |
ENSMUST00000030785.8
ENSMUST00000105658.1 ENSMUST00000105659.2 |
Espn
|
espin |
| chr7_-_119793958 | 1.13 |
ENSMUST00000106523.1
ENSMUST00000063902.7 ENSMUST00000150844.1 |
Eri2
|
exoribonuclease 2 |
| chr12_-_36156781 | 1.12 |
ENSMUST00000020856.4
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr1_+_66386968 | 1.10 |
ENSMUST00000145419.1
|
Map2
|
microtubule-associated protein 2 |
| chr7_+_5056856 | 1.10 |
ENSMUST00000131368.1
ENSMUST00000123956.1 |
Ccdc106
|
coiled-coil domain containing 106 |
| chr15_-_83149307 | 1.09 |
ENSMUST00000100375.4
|
Poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr5_-_112356033 | 1.08 |
ENSMUST00000051117.2
|
Gm6583
|
predicted gene 6583 |
| chr2_+_31759932 | 1.08 |
ENSMUST00000028190.6
|
Abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr6_-_56923927 | 1.07 |
ENSMUST00000031793.5
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr10_-_81001338 | 1.06 |
ENSMUST00000099462.1
ENSMUST00000118233.1 |
Gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr15_+_85017138 | 1.05 |
ENSMUST00000023070.5
|
Upk3a
|
uroplakin 3A |
| chr9_-_66124872 | 1.03 |
ENSMUST00000034946.8
|
Snx1
|
sorting nexin 1 |
| chr8_+_84872105 | 0.97 |
ENSMUST00000136026.1
ENSMUST00000170296.1 |
Syce2
|
synaptonemal complex central element protein 2 |
| chr9_+_57940104 | 0.95 |
ENSMUST00000043059.7
|
Sema7a
|
sema domain, immunoglobulin domain (Ig), and GPI membrane anchor, (semaphorin) 7A |
| chr15_-_93519499 | 0.82 |
ENSMUST00000109255.2
|
Prickle1
|
prickle homolog 1 (Drosophila) |
| chr12_+_111758923 | 0.72 |
ENSMUST00000118471.1
ENSMUST00000122300.1 |
Klc1
|
kinesin light chain 1 |
| chr1_+_179668202 | 0.68 |
ENSMUST00000040538.3
|
Sccpdh
|
saccharopine dehydrogenase (putative) |
| chr8_+_106210936 | 0.65 |
ENSMUST00000071592.5
|
Prmt7
|
protein arginine N-methyltransferase 7 |
| chr12_+_111758848 | 0.64 |
ENSMUST00000084941.5
|
Klc1
|
kinesin light chain 1 |
| chr5_+_57718021 | 0.61 |
ENSMUST00000094783.3
ENSMUST00000068110.7 |
Pcdh7
|
protocadherin 7 |
| chr12_+_33957645 | 0.58 |
ENSMUST00000049089.5
|
Twist1
|
twist basic helix-loop-helix transcription factor 1 |
| chr17_+_71673255 | 0.58 |
ENSMUST00000097284.3
|
Fam179a
|
family with sequence similarity 179, member A |
| chr17_+_47688992 | 0.57 |
ENSMUST00000156118.1
|
Frs3
|
fibroblast growth factor receptor substrate 3 |
| chr3_-_94412883 | 0.56 |
ENSMUST00000181305.1
|
1700040D17Rik
|
RIKEN cDNA 1700040D17 gene |
| chr4_+_134343181 | 0.55 |
ENSMUST00000105873.1
ENSMUST00000105874.2 |
Slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr16_-_91597636 | 0.53 |
ENSMUST00000023686.8
|
Tmem50b
|
transmembrane protein 50B |
| chr2_+_31759993 | 0.53 |
ENSMUST00000124089.1
|
Abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr5_-_141000590 | 0.49 |
ENSMUST00000085786.5
|
Card11
|
caspase recruitment domain family, member 11 |
| chr8_+_106211016 | 0.49 |
ENSMUST00000109297.1
|
Prmt7
|
protein arginine N-methyltransferase 7 |
| chr1_+_128103297 | 0.34 |
ENSMUST00000036288.4
|
R3hdm1
|
R3H domain containing 1 |
| chr2_+_150323702 | 0.33 |
ENSMUST00000133235.2
|
Gm10130
|
predicted gene 10130 |
| chr2_-_119787504 | 0.32 |
ENSMUST00000110793.1
ENSMUST00000099529.2 ENSMUST00000048493.5 |
Rpap1
|
RNA polymerase II associated protein 1 |
| chr8_+_92827273 | 0.31 |
ENSMUST00000034187.7
|
Mmp2
|
matrix metallopeptidase 2 |
| chr9_-_112187898 | 0.30 |
ENSMUST00000178410.1
ENSMUST00000172380.3 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr9_+_108490676 | 0.30 |
ENSMUST00000178075.1
ENSMUST00000085044.7 ENSMUST00000166103.2 ENSMUST00000006854.7 |
Usp19
|
ubiquitin specific peptidase 19 |
| chr4_+_134343466 | 0.26 |
ENSMUST00000105872.1
|
Slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr17_-_83631892 | 0.23 |
ENSMUST00000051482.1
|
Kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr14_-_26170283 | 0.23 |
ENSMUST00000100809.4
|
Plac9b
|
placenta specific 9b |
| chrX_-_79671434 | 0.21 |
ENSMUST00000052283.6
|
Mageb16
|
melanoma antigen family B, 16 |
| chr11_+_4895328 | 0.17 |
ENSMUST00000038237.1
|
Thoc5
|
THO complex 5 |
| chr8_-_70776650 | 0.16 |
ENSMUST00000034296.8
|
Pik3r2
|
phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 2 (p85 beta) |
| chr1_-_166002591 | 0.15 |
ENSMUST00000111429.4
ENSMUST00000176800.1 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr18_-_84589491 | 0.14 |
ENSMUST00000125763.1
|
Zfp407
|
zinc finger protein 407 |
| chr10_+_81136223 | 0.13 |
ENSMUST00000048128.8
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr11_+_4895316 | 0.11 |
ENSMUST00000101615.2
|
Thoc5
|
THO complex 5 |
| chr16_+_18348181 | 0.11 |
ENSMUST00000115614.2
ENSMUST00000115613.1 ENSMUST00000090103.3 |
Arvcf
|
armadillo repeat gene deleted in velo-cardio-facial syndrome |
| chr11_-_5898771 | 0.07 |
ENSMUST00000102921.3
|
Myl7
|
myosin, light polypeptide 7, regulatory |
| chr5_-_125341043 | 0.01 |
ENSMUST00000111390.1
ENSMUST00000086075.6 |
Scarb1
|
scavenger receptor class B, member 1 |
| chr18_+_11839220 | 0.01 |
ENSMUST00000171109.1
ENSMUST00000046948.8 |
Cables1
|
CDK5 and Abl enzyme substrate 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mtf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 42.4 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 1.7 | 13.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 1.3 | 9.2 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 1.1 | 3.3 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 1.0 | 3.0 | GO:0030862 | positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.9 | 5.4 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.7 | 2.7 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.5 | 1.6 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) activation of protein kinase C activity(GO:1990051) |
| 0.4 | 1.3 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.3 | 0.8 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.3 | 1.4 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.3 | 0.8 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 0.2 | 4.8 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 1.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.2 | 4.0 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.2 | 8.1 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.2 | 0.6 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 0.2 | 1.1 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.2 | 1.1 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.2 | 1.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.1 | 1.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 1.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.5 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.1 | 0.9 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 1.0 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 1.4 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.3 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 1.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 1.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.5 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.0 | 3.6 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.3 | GO:2000002 | regulation of mRNA export from nucleus(GO:0010793) negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 1.0 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 3.9 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 1.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 2.3 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 1.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 1.9 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 | 0.3 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 1.8 | GO:0098656 | anion transmembrane transport(GO:0098656) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 8.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.5 | 9.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 1.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.3 | 1.0 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.2 | 3.6 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 1.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 3.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 4.8 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 1.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 1.0 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 2.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.4 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 1.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 5.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 3.3 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 9.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 12.2 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.4 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.9 | 13.2 | GO:0019841 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.4 | 2.7 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 9.2 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.3 | 1.5 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.3 | 1.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.3 | 1.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.3 | 1.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.2 | 15.5 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.2 | 2.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 1.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 4.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 3.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 3.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 4.4 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 1.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 19.7 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 14.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 4.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 3.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 3.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 4.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 13.2 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.3 | 12.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 8.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.5 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 1.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 4.0 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 1.3 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.6 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |