Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Msx2_Hoxd4
Z-value: 1.33
Transcription factors associated with Msx2_Hoxd4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
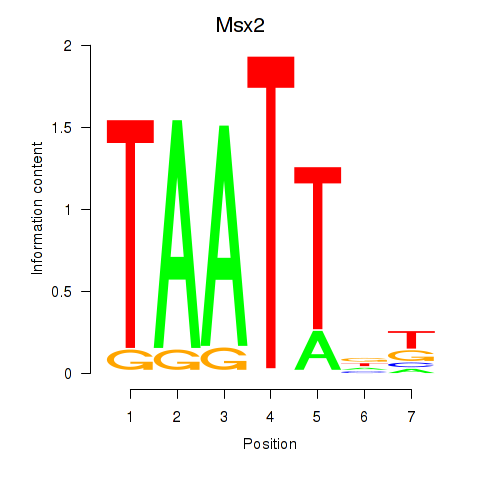
Msx2
|
ENSMUSG00000021469.8 | msh homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Msx2 | mm10_v2_chr13_-_53473074_53473074 | 0.40 | 1.7e-02 | Click! |
Activity profile of Msx2_Hoxd4 motif
Sorted Z-values of Msx2_Hoxd4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_79285502 | 10.45 |
ENSMUST00000165408.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr15_-_79285470 | 7.47 |
ENSMUST00000170955.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr15_-_36879816 | 5.60 |
ENSMUST00000100713.2
|
Gm10384
|
predicted gene 10384 |
| chr2_+_4017727 | 4.90 |
ENSMUST00000177457.1
|
Frmd4a
|
FERM domain containing 4A |
| chrX_+_21484532 | 4.69 |
ENSMUST00000089188.2
|
Agtr2
|
angiotensin II receptor, type 2 |
| chr6_-_36811361 | 4.19 |
ENSMUST00000101534.1
|
Ptn
|
pleiotrophin |
| chr1_-_132390301 | 4.18 |
ENSMUST00000132435.1
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr11_+_95010277 | 3.61 |
ENSMUST00000124735.1
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr11_+_67171095 | 2.83 |
ENSMUST00000018641.7
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr3_+_40800778 | 2.74 |
ENSMUST00000169566.1
|
Plk4
|
polo-like kinase 4 |
| chr3_-_150073620 | 2.67 |
ENSMUST00000057740.5
|
Rpsa-ps10
|
ribosomal protein SA, pseudogene 10 |
| chrX_+_56454871 | 2.61 |
ENSMUST00000039374.2
ENSMUST00000101553.2 |
Ddx26b
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr11_+_67171027 | 2.59 |
ENSMUST00000170159.1
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr3_-_36571952 | 2.27 |
ENSMUST00000029270.3
|
Ccna2
|
cyclin A2 |
| chr18_+_34840575 | 2.18 |
ENSMUST00000043484.7
|
Reep2
|
receptor accessory protein 2 |
| chr19_-_46044914 | 2.15 |
ENSMUST00000026252.7
|
Ldb1
|
LIM domain binding 1 |
| chrX_+_159303266 | 1.91 |
ENSMUST00000112491.1
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr3_+_68869563 | 1.88 |
ENSMUST00000054551.2
|
1110032F04Rik
|
RIKEN cDNA 1110032F04 gene |
| chr1_-_163289214 | 1.85 |
ENSMUST00000183691.1
|
Prrx1
|
paired related homeobox 1 |
| chr2_+_109280738 | 1.81 |
ENSMUST00000028527.7
|
Kif18a
|
kinesin family member 18A |
| chrX_+_106583184 | 1.73 |
ENSMUST00000101296.2
ENSMUST00000101297.3 |
Gm5127
|
predicted gene 5127 |
| chrX_-_162964557 | 1.71 |
ENSMUST00000038769.2
|
S100g
|
S100 calcium binding protein G |
| chrX_+_136245065 | 1.66 |
ENSMUST00000048687.4
|
Wbp5
|
WW domain binding protein 5 |
| chrX_-_74246534 | 1.63 |
ENSMUST00000101454.2
ENSMUST00000033699.6 |
Flna
|
filamin, alpha |
| chr3_-_116253467 | 1.62 |
ENSMUST00000090473.5
|
Gpr88
|
G-protein coupled receptor 88 |
| chr16_-_58524164 | 1.58 |
ENSMUST00000126978.1
ENSMUST00000123918.1 |
St3gal6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chrX_+_159708593 | 1.52 |
ENSMUST00000080394.6
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr6_-_136941887 | 1.48 |
ENSMUST00000111891.1
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr7_-_115824699 | 1.42 |
ENSMUST00000169129.1
|
Sox6
|
SRY-box containing gene 6 |
| chr4_-_141078302 | 1.41 |
ENSMUST00000030760.8
|
Necap2
|
NECAP endocytosis associated 2 |
| chr6_-_87533219 | 1.36 |
ENSMUST00000113637.2
ENSMUST00000071024.6 |
Arhgap25
|
Rho GTPase activating protein 25 |
| chr18_+_44104407 | 1.35 |
ENSMUST00000081271.5
|
Spink12
|
serine peptidase inhibitor, Kazal type 11 |
| chr3_-_14778452 | 1.32 |
ENSMUST00000094365.4
|
Car1
|
carbonic anhydrase 1 |
| chr2_-_72986716 | 1.31 |
ENSMUST00000112062.1
|
Gm11084
|
predicted gene 11084 |
| chr15_-_101694299 | 1.29 |
ENSMUST00000023788.6
|
Krt6a
|
keratin 6A |
| chr7_-_121074501 | 1.29 |
ENSMUST00000047194.2
|
Igsf6
|
immunoglobulin superfamily, member 6 |
| chr10_-_63203930 | 1.26 |
ENSMUST00000095580.2
|
Mypn
|
myopalladin |
| chrX_-_150814265 | 1.26 |
ENSMUST00000026302.6
ENSMUST00000129768.1 ENSMUST00000112699.2 |
Maged2
|
melanoma antigen, family D, 2 |
| chr1_+_63176818 | 1.17 |
ENSMUST00000129339.1
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr2_-_73453918 | 1.13 |
ENSMUST00000102679.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr9_-_77544870 | 1.13 |
ENSMUST00000183873.1
|
Lrrc1
|
leucine rich repeat containing 1 |
| chr13_+_23544052 | 1.13 |
ENSMUST00000075558.2
|
Hist1h3f
|
histone cluster 1, H3f |
| chr13_+_49544443 | 1.13 |
ENSMUST00000177948.1
ENSMUST00000021820.6 |
Aspn
|
asporin |
| chr3_-_75270073 | 1.12 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr11_-_109472611 | 1.11 |
ENSMUST00000168740.1
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr7_+_144838590 | 1.10 |
ENSMUST00000105898.1
|
Fgf3
|
fibroblast growth factor 3 |
| chrX_+_150547375 | 1.08 |
ENSMUST00000066337.6
ENSMUST00000112715.1 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr4_-_129121889 | 1.07 |
ENSMUST00000139450.1
ENSMUST00000125931.1 ENSMUST00000116444.2 |
Hpca
|
hippocalcin |
| chr2_-_164389095 | 1.05 |
ENSMUST00000167427.1
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr10_+_14523062 | 1.01 |
ENSMUST00000096020.5
|
Gm10335
|
predicted gene 10335 |
| chr9_-_77544829 | 1.01 |
ENSMUST00000183734.1
|
Lrrc1
|
leucine rich repeat containing 1 |
| chr4_-_149126688 | 0.99 |
ENSMUST00000030815.2
|
Cort
|
cortistatin |
| chr8_-_8639363 | 0.99 |
ENSMUST00000152698.1
|
Efnb2
|
ephrin B2 |
| chr1_-_174250976 | 0.97 |
ENSMUST00000061990.4
|
Olfr419
|
olfactory receptor 419 |
| chr7_-_45921418 | 0.96 |
ENSMUST00000038876.5
|
Emp3
|
epithelial membrane protein 3 |
| chr14_+_115042752 | 0.95 |
ENSMUST00000134140.2
|
Mir17hg
|
Mir17 host gene 1 (non-protein coding) |
| chr12_+_36314160 | 0.94 |
ENSMUST00000041407.5
|
Sostdc1
|
sclerostin domain containing 1 |
| chrX_-_164258186 | 0.94 |
ENSMUST00000112265.2
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr4_-_3872105 | 0.93 |
ENSMUST00000105158.1
|
Mos
|
Moloney sarcoma oncogene |
| chr4_+_101507947 | 0.93 |
ENSMUST00000149047.1
ENSMUST00000106929.3 |
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr3_+_64081642 | 0.91 |
ENSMUST00000029406.4
|
Vmn2r1
|
vomeronasal 2, receptor 1 |
| chr17_+_46161021 | 0.89 |
ENSMUST00000024748.7
ENSMUST00000172170.1 |
Gtpbp2
|
GTP binding protein 2 |
| chr13_+_75967704 | 0.87 |
ENSMUST00000022081.1
|
Spata9
|
spermatogenesis associated 9 |
| chr14_+_53795455 | 0.85 |
ENSMUST00000103671.2
|
B230359F08Rik
|
RIKEN cDNA B230359F08 gene |
| chr2_-_45110336 | 0.82 |
ENSMUST00000028229.6
ENSMUST00000152232.1 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr3_-_102964124 | 0.82 |
ENSMUST00000058899.8
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr12_+_38783503 | 0.80 |
ENSMUST00000159334.1
|
Etv1
|
ets variant gene 1 |
| chr11_+_103133333 | 0.80 |
ENSMUST00000124928.1
ENSMUST00000062530.4 |
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr6_+_136518820 | 0.79 |
ENSMUST00000032335.6
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr7_+_126950518 | 0.77 |
ENSMUST00000106335.1
ENSMUST00000146017.1 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chr19_-_53371766 | 0.76 |
ENSMUST00000086887.1
|
Gm10197
|
predicted gene 10197 |
| chr7_-_45920830 | 0.75 |
ENSMUST00000164119.1
|
Emp3
|
epithelial membrane protein 3 |
| chr2_-_33428222 | 0.73 |
ENSMUST00000091037.2
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chr2_-_45112890 | 0.73 |
ENSMUST00000076836.6
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr2_+_80638798 | 0.72 |
ENSMUST00000028382.6
ENSMUST00000124377.1 |
Nup35
|
nucleoporin 35 |
| chr6_-_131316398 | 0.72 |
ENSMUST00000121078.1
|
Styk1
|
serine/threonine/tyrosine kinase 1 |
| chr1_+_136683375 | 0.72 |
ENSMUST00000181524.1
|
Gm19705
|
predicted gene, 19705 |
| chr18_+_68337504 | 0.71 |
ENSMUST00000172148.1
|
Mc5r
|
melanocortin 5 receptor |
| chr1_+_186749368 | 0.71 |
ENSMUST00000180869.1
|
A430105J06Rik
|
RIKEN cDNA A430105J06 gene |
| chr1_+_191025350 | 0.70 |
ENSMUST00000181050.1
|
A230020J21Rik
|
RIKEN cDNA A230020J21 gene |
| chr9_-_112187766 | 0.70 |
ENSMUST00000111872.2
ENSMUST00000164754.2 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr1_-_149961230 | 0.66 |
ENSMUST00000070200.8
|
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr17_-_14694223 | 0.64 |
ENSMUST00000170872.1
|
Thbs2
|
thrombospondin 2 |
| chr10_-_13324160 | 0.63 |
ENSMUST00000105545.4
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr19_+_46396885 | 0.61 |
ENSMUST00000039922.6
ENSMUST00000111867.2 ENSMUST00000120778.1 |
Sufu
|
suppressor of fused homolog (Drosophila) |
| chr11_+_103133303 | 0.59 |
ENSMUST00000107037.1
|
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr13_+_23555023 | 0.59 |
ENSMUST00000045301.6
|
Hist1h1d
|
histone cluster 1, H1d |
| chr17_-_24073479 | 0.59 |
ENSMUST00000017090.5
|
Kctd5
|
potassium channel tetramerisation domain containing 5 |
| chr11_-_87826023 | 0.58 |
ENSMUST00000103177.3
|
Lpo
|
lactoperoxidase |
| chr15_+_85510812 | 0.57 |
ENSMUST00000079690.2
|
Gm4825
|
predicted pseudogene 4825 |
| chr1_-_183147461 | 0.56 |
ENSMUST00000171366.1
|
Disp1
|
dispatched homolog 1 (Drosophila) |
| chr5_-_53707532 | 0.56 |
ENSMUST00000031093.3
|
Cckar
|
cholecystokinin A receptor |
| chr18_+_37518341 | 0.55 |
ENSMUST00000097609.1
|
Pcdhb22
|
protocadherin beta 22 |
| chr18_-_43477764 | 0.52 |
ENSMUST00000057110.9
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr4_-_73950834 | 0.51 |
ENSMUST00000095023.1
ENSMUST00000030101.3 |
2310002L09Rik
|
RIKEN cDNA 2310002L09 gene |
| chr2_+_155751117 | 0.51 |
ENSMUST00000029140.5
ENSMUST00000132608.1 |
Procr
|
protein C receptor, endothelial |
| chr2_-_152580300 | 0.50 |
ENSMUST00000053180.3
|
Defb19
|
defensin beta 19 |
| chr1_-_152625212 | 0.50 |
ENSMUST00000027760.7
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr4_-_82850721 | 0.50 |
ENSMUST00000139401.1
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr1_-_150164943 | 0.49 |
ENSMUST00000181308.1
|
Gm26687
|
predicted gene, 26687 |
| chrX_-_94123359 | 0.49 |
ENSMUST00000137853.1
ENSMUST00000088102.5 ENSMUST00000113927.1 |
Zfx
|
zinc finger protein X-linked |
| chr9_-_114390633 | 0.48 |
ENSMUST00000084881.4
|
Crtap
|
cartilage associated protein |
| chr2_-_60125651 | 0.47 |
ENSMUST00000112550.1
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr2_+_57997884 | 0.46 |
ENSMUST00000112616.1
ENSMUST00000166729.1 |
Galnt5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 |
| chr9_-_107770945 | 0.46 |
ENSMUST00000183248.1
ENSMUST00000182022.1 ENSMUST00000035199.6 ENSMUST00000182659.1 |
Rbm5
|
RNA binding motif protein 5 |
| chr4_+_3940747 | 0.46 |
ENSMUST00000119403.1
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr19_-_55241236 | 0.46 |
ENSMUST00000069183.6
|
Gucy2g
|
guanylate cyclase 2g |
| chr4_-_119492563 | 0.44 |
ENSMUST00000049994.7
|
Rimkla
|
ribosomal modification protein rimK-like family member A |
| chr7_+_126950687 | 0.44 |
ENSMUST00000106333.1
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr3_-_49757257 | 0.44 |
ENSMUST00000035931.7
|
Pcdh18
|
protocadherin 18 |
| chr2_+_76650264 | 0.43 |
ENSMUST00000099986.2
|
Dfnb59
|
deafness, autosomal recessive 59 (human) |
| chr2_-_169962987 | 0.43 |
ENSMUST00000087964.2
|
AY702102
|
cDNA sequence AY702102 |
| chr4_-_99829180 | 0.41 |
ENSMUST00000146258.1
|
Itgb3bp
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr4_+_101507855 | 0.40 |
ENSMUST00000038207.5
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr15_+_21111452 | 0.40 |
ENSMUST00000075132.6
|
Cdh12
|
cadherin 12 |
| chr5_+_48242549 | 0.40 |
ENSMUST00000172493.1
|
Slit2
|
slit homolog 2 (Drosophila) |
| chr18_+_52767994 | 0.38 |
ENSMUST00000025413.7
ENSMUST00000163742.2 ENSMUST00000178011.1 |
Sncaip
|
synuclein, alpha interacting protein (synphilin) |
| chr9_+_30942541 | 0.38 |
ENSMUST00000068135.6
|
Adamts8
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 8 |
| chr1_+_159737510 | 0.38 |
ENSMUST00000111669.3
|
Tnr
|
tenascin R |
| chr5_-_62765618 | 0.38 |
ENSMUST00000159470.1
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr3_-_66296807 | 0.37 |
ENSMUST00000029419.7
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr2_-_72813665 | 0.37 |
ENSMUST00000136807.1
ENSMUST00000148327.1 |
6430710C18Rik
|
RIKEN cDNA 6430710C18 gene |
| chrX_+_133850980 | 0.37 |
ENSMUST00000033602.8
|
Tnmd
|
tenomodulin |
| chr6_-_124779686 | 0.36 |
ENSMUST00000147669.1
ENSMUST00000128697.1 ENSMUST00000032218.3 ENSMUST00000112475.2 |
Lrrc23
|
leucine rich repeat containing 23 |
| chr5_-_31179901 | 0.35 |
ENSMUST00000101411.2
ENSMUST00000140793.1 |
Gtf3c2
|
general transcription factor IIIC, polypeptide 2, beta |
| chr5_-_72168142 | 0.34 |
ENSMUST00000013693.6
|
Commd8
|
COMM domain containing 8 |
| chr8_-_120228221 | 0.34 |
ENSMUST00000183235.1
|
A330074K22Rik
|
RIKEN cDNA A330074K22 gene |
| chr13_-_102906046 | 0.34 |
ENSMUST00000171791.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_-_133661318 | 0.33 |
ENSMUST00000179598.1
ENSMUST00000027736.6 |
Zbed6
Zc3h11a
|
zinc finger, BED domain containing 6 zinc finger CCCH type containing 11A |
| chr1_+_171840557 | 0.32 |
ENSMUST00000135386.1
|
Cd84
|
CD84 antigen |
| chr17_+_17402672 | 0.32 |
ENSMUST00000115576.2
|
Lix1
|
limb expression 1 homolog (chicken) |
| chr1_-_78968079 | 0.32 |
ENSMUST00000049117.5
|
Gm5830
|
predicted pseudogene 5830 |
| chr4_-_62519885 | 0.32 |
ENSMUST00000107444.1
ENSMUST00000030090.3 |
Alad
|
aminolevulinate, delta-, dehydratase |
| chr10_+_115569986 | 0.31 |
ENSMUST00000173620.1
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr8_+_81856324 | 0.31 |
ENSMUST00000109851.2
|
Inpp4b
|
inositol polyphosphate-4-phosphatase, type II |
| chr9_-_118014160 | 0.31 |
ENSMUST00000111769.3
|
Zcwpw2
|
zinc finger, CW type with PWWP domain 2 |
| chr7_+_126950837 | 0.31 |
ENSMUST00000106332.1
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr1_+_106938953 | 0.30 |
ENSMUST00000112724.2
|
Serpinb12
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr13_-_75943812 | 0.30 |
ENSMUST00000022078.5
ENSMUST00000109606.1 |
Rhobtb3
|
Rho-related BTB domain containing 3 |
| chr3_+_84952146 | 0.29 |
ENSMUST00000029727.7
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr1_+_70725902 | 0.29 |
ENSMUST00000161937.1
ENSMUST00000162182.1 |
Vwc2l
|
von Willebrand factor C domain-containing protein 2-like |
| chr2_-_163417092 | 0.29 |
ENSMUST00000127038.1
|
Oser1
|
oxidative stress responsive serine rich 1 |
| chr10_+_128303322 | 0.28 |
ENSMUST00000005825.6
|
Pan2
|
PAN2 polyA specific ribonuclease subunit homolog (S. cerevisiae) |
| chr13_+_93304799 | 0.26 |
ENSMUST00000080127.5
|
Homer1
|
homer homolog 1 (Drosophila) |
| chr2_-_33086366 | 0.26 |
ENSMUST00000049618.2
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr14_-_88471396 | 0.26 |
ENSMUST00000061628.5
|
Pcdh20
|
protocadherin 20 |
| chr18_+_59062462 | 0.24 |
ENSMUST00000058633.2
ENSMUST00000175897.1 ENSMUST00000118510.1 ENSMUST00000175830.1 |
A730017C20Rik
|
RIKEN cDNA A730017C20 gene |
| chr18_+_37320374 | 0.24 |
ENSMUST00000078271.2
|
Pcdhb5
|
protocadherin beta 5 |
| chr6_-_67491849 | 0.23 |
ENSMUST00000118364.1
|
Il23r
|
interleukin 23 receptor |
| chr5_-_31180110 | 0.22 |
ENSMUST00000043161.6
ENSMUST00000088010.5 |
Gtf3c2
|
general transcription factor IIIC, polypeptide 2, beta |
| chr5_-_151586924 | 0.22 |
ENSMUST00000165928.1
|
Vmn2r18
|
vomeronasal 2, receptor 18 |
| chrX_+_101640056 | 0.22 |
ENSMUST00000119299.1
ENSMUST00000044475.4 |
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) |
| chr11_+_73350839 | 0.22 |
ENSMUST00000120137.1
|
Olfr20
|
olfactory receptor 20 |
| chr7_+_123123870 | 0.22 |
ENSMUST00000094053.5
|
Tnrc6a
|
trinucleotide repeat containing 6a |
| chr1_+_70725715 | 0.21 |
ENSMUST00000053922.5
|
Vwc2l
|
von Willebrand factor C domain-containing protein 2-like |
| chr5_+_87925579 | 0.21 |
ENSMUST00000001667.6
ENSMUST00000113267.1 |
Csn3
|
casein kappa |
| chr12_+_108179738 | 0.20 |
ENSMUST00000101055.4
|
Ccnk
|
cyclin K |
| chr6_-_119963733 | 0.19 |
ENSMUST00000161512.2
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr8_+_31111816 | 0.18 |
ENSMUST00000046941.7
|
Rnf122
|
ring finger protein 122 |
| chr2_+_36452587 | 0.17 |
ENSMUST00000072854.1
|
Olfr340
|
olfactory receptor 340 |
| chr13_+_51408618 | 0.16 |
ENSMUST00000087978.3
|
S1pr3
|
sphingosine-1-phosphate receptor 3 |
| chr9_+_38877126 | 0.16 |
ENSMUST00000078289.2
|
Olfr926
|
olfactory receptor 926 |
| chr2_+_74691090 | 0.16 |
ENSMUST00000061745.3
|
Hoxd10
|
homeobox D10 |
| chr4_-_73790602 | 0.16 |
ENSMUST00000058292.6
ENSMUST00000102837.2 |
Rasef
|
RAS and EF hand domain containing |
| chr11_-_74724670 | 0.16 |
ENSMUST00000021091.8
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr4_+_35152056 | 0.16 |
ENSMUST00000058595.6
|
Ifnk
|
interferon kappa |
| chr18_+_4920509 | 0.15 |
ENSMUST00000126977.1
|
Svil
|
supervillin |
| chr1_+_180109192 | 0.14 |
ENSMUST00000143176.1
ENSMUST00000135056.1 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr2_+_180710117 | 0.14 |
ENSMUST00000029090.2
|
Gid8
|
GID complex subunit 8 homolog (S. cerevisiae) |
| chr1_+_72284367 | 0.14 |
ENSMUST00000027380.5
ENSMUST00000141783.1 |
Tmem169
|
transmembrane protein 169 |
| chr12_+_52516077 | 0.13 |
ENSMUST00000110725.1
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr1_-_186749304 | 0.13 |
ENSMUST00000001339.5
|
Rrp15
|
ribosomal RNA processing 15 homolog (S. cerevisiae) |
| chr3_+_59952185 | 0.13 |
ENSMUST00000094227.3
|
Gm9696
|
predicted gene 9696 |
| chr11_+_96292453 | 0.12 |
ENSMUST00000173432.1
|
Hoxb6
|
homeobox B6 |
| chr10_-_14718191 | 0.12 |
ENSMUST00000020016.4
|
Gje1
|
gap junction protein, epsilon 1 |
| chr9_+_64235201 | 0.12 |
ENSMUST00000039011.3
|
Uchl4
|
ubiquitin carboxyl-terminal esterase L4 |
| chr7_-_38019505 | 0.11 |
ENSMUST00000085513.4
|
Uri1
|
URI1, prefoldin-like chaperone |
| chr19_-_39740999 | 0.10 |
ENSMUST00000099472.3
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68 |
| chr1_+_137928100 | 0.10 |
ENSMUST00000054333.2
|
A130050O07Rik
|
RIKEN cDNA A130050O07 gene |
| chr10_+_5593718 | 0.10 |
ENSMUST00000051809.8
|
Myct1
|
myc target 1 |
| chr10_-_129160982 | 0.09 |
ENSMUST00000078914.2
|
Olfr771
|
olfactory receptor 771 |
| chr5_-_23616528 | 0.09 |
ENSMUST00000088392.4
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr18_-_37969742 | 0.08 |
ENSMUST00000166148.1
ENSMUST00000163131.1 ENSMUST00000043437.7 |
Fchsd1
|
FCH and double SH3 domains 1 |
| chr13_+_93304066 | 0.08 |
ENSMUST00000109493.1
|
Homer1
|
homer homolog 1 (Drosophila) |
| chr3_+_106113229 | 0.07 |
ENSMUST00000079132.5
ENSMUST00000139086.1 |
Chia
|
chitinase, acidic |
| chr11_-_99322943 | 0.06 |
ENSMUST00000038004.2
|
Krt25
|
keratin 25 |
| chr7_+_84853573 | 0.05 |
ENSMUST00000078172.4
|
Olfr291
|
olfactory receptor 291 |
| chr12_-_27160498 | 0.04 |
ENSMUST00000182592.1
|
Gm9866
|
predicted gene 9866 |
| chr6_-_23650206 | 0.04 |
ENSMUST00000115354.1
|
Rnf133
|
ring finger protein 133 |
| chr14_-_52036143 | 0.04 |
ENSMUST00000052560.4
|
Olfr221
|
olfactory receptor 221 |
| chr9_-_89705017 | 0.02 |
ENSMUST00000058488.6
|
Tmed3
|
transmembrane emp24 domain containing 3 |
| chr15_+_92597104 | 0.01 |
ENSMUST00000035399.8
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr11_+_98798653 | 0.01 |
ENSMUST00000037930.6
|
Msl1
|
male-specific lethal 1 homolog (Drosophila) |
| chr17_-_35046539 | 0.01 |
ENSMUST00000007250.7
|
Msh5
|
mutS homolog 5 (E. coli) |
| chr12_-_27160311 | 0.00 |
ENSMUST00000182473.1
ENSMUST00000177636.1 ENSMUST00000183238.1 |
Gm9866
|
predicted gene 9866 |
| chr3_+_116968267 | 0.00 |
ENSMUST00000117592.1
|
4930455H04Rik
|
RIKEN cDNA 4930455H04 gene |
| chr13_+_76098734 | 0.00 |
ENSMUST00000091466.3
|
Ttc37
|
tetratricopeptide repeat domain 37 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Msx2_Hoxd4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 1.0 | 4.2 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.8 | 17.9 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.5 | 1.6 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.5 | 2.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.4 | 1.6 | GO:0061743 | motor learning(GO:0061743) |
| 0.4 | 2.7 | GO:0098535 | positive regulation of centriole replication(GO:0046601) de novo centriole assembly(GO:0098535) |
| 0.4 | 2.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.4 | 5.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.4 | 1.1 | GO:0031283 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.3 | 0.9 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.3 | 0.9 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.3 | 1.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 1.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.2 | 0.9 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.2 | 1.3 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.2 | 0.6 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.2 | 1.5 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.2 | 1.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.4 | GO:0090260 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.1 | 1.4 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.4 | GO:0048690 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 | 1.0 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 1.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 1.9 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.7 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 4.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.3 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.1 | 1.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.6 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 1.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 1.1 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.4 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 1.0 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 1.5 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 1.8 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 4.0 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 0.9 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.3 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.2 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 1.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.5 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 2.3 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.8 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.2 | GO:0043622 | cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) |
| 0.0 | 1.4 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.7 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 1.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.6 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 1.6 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 1.0 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 1.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.6 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 1.1 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.9 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.4 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 1.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.5 | GO:0060746 | parental behavior(GO:0060746) |
| 0.0 | 0.7 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.3 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 | 1.9 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.2 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.3 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.2 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.0 | 0.2 | GO:1901550 | regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.0 | 0.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.4 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.7 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 17.9 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.7 | 5.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.5 | 2.7 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 1.6 | GO:0031523 | Myb complex(GO:0031523) |
| 0.3 | 2.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.3 | 1.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.2 | 1.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.4 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 0.7 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 1.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 0.6 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.2 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.1 | 1.4 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 4.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 2.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.6 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 4.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 1.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.2 | 4.7 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.4 | 1.6 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.4 | 1.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.3 | 1.6 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.2 | 1.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 1.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 2.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 1.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 1.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.7 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 1.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.9 | GO:0036122 | BMP binding(GO:0036122) BMP receptor activity(GO:0098821) |
| 0.1 | 0.5 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.4 | GO:0048495 | GTPase inhibitor activity(GO:0005095) Roundabout binding(GO:0048495) |
| 0.0 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 2.5 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 1.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 1.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.6 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 1.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 15.2 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 1.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 4.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.4 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 3.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 4.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.6 | GO:0004601 | peroxidase activity(GO:0004601) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 6.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 3.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.9 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.1 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 1.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 2.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.7 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 6.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 1.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 2.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |