Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Mnt
Z-value: 1.10
Transcription factors associated with Mnt
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mnt
|
ENSMUSG00000000282.6 | max binding protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mnt | mm10_v2_chr11_+_74830920_74831005 | 0.16 | 3.6e-01 | Click! |
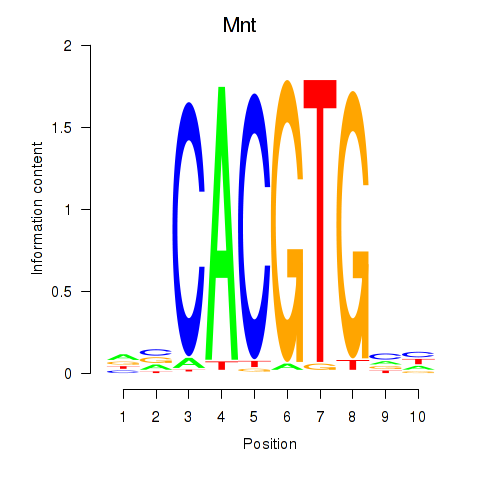
Activity profile of Mnt motif
Sorted Z-values of Mnt motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_80671829 | 2.42 |
ENSMUST00000023044.5
|
Fam83f
|
family with sequence similarity 83, member F |
| chr16_+_32608973 | 1.84 |
ENSMUST00000120680.1
|
Tfrc
|
transferrin receptor |
| chrX_-_136068236 | 1.69 |
ENSMUST00000049130.7
|
Bex2
|
brain expressed X-linked 2 |
| chrX_-_136215443 | 1.52 |
ENSMUST00000113120.1
ENSMUST00000113118.1 ENSMUST00000058125.8 |
Bex1
|
brain expressed gene 1 |
| chrX_+_136138996 | 1.50 |
ENSMUST00000116527.1
|
Bex4
|
brain expressed gene 4 |
| chr17_-_56476462 | 1.49 |
ENSMUST00000067538.5
|
Ptprs
|
protein tyrosine phosphatase, receptor type, S |
| chrX_+_153139941 | 1.47 |
ENSMUST00000039720.4
ENSMUST00000144175.2 |
Rragb
|
Ras-related GTP binding B |
| chr1_+_172481788 | 1.44 |
ENSMUST00000127052.1
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr5_-_76951560 | 1.42 |
ENSMUST00000140076.1
|
Ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr11_+_69095217 | 1.38 |
ENSMUST00000101004.2
|
Per1
|
period circadian clock 1 |
| chr9_-_22389113 | 1.34 |
ENSMUST00000040912.7
|
Anln
|
anillin, actin binding protein |
| chr1_+_172482199 | 1.32 |
ENSMUST00000135267.1
ENSMUST00000052629.6 ENSMUST00000111235.2 |
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr12_+_102129019 | 1.27 |
ENSMUST00000079020.4
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr15_+_81811414 | 1.26 |
ENSMUST00000023024.7
|
Tef
|
thyrotroph embryonic factor |
| chr1_-_119422239 | 1.22 |
ENSMUST00000038765.5
|
Inhbb
|
inhibin beta-B |
| chr15_-_44788016 | 1.18 |
ENSMUST00000090057.4
ENSMUST00000110269.1 |
Sybu
|
syntabulin (syntaxin-interacting) |
| chr17_-_35000848 | 1.11 |
ENSMUST00000166828.3
|
D17H6S56E-5
|
DNA segment, Chr 17, human D6S56E 5 |
| chrX_+_50841434 | 1.11 |
ENSMUST00000114887.2
|
2610018G03Rik
|
RIKEN cDNA 2610018G03 gene |
| chrX_-_136203637 | 1.08 |
ENSMUST00000151592.1
ENSMUST00000131510.1 ENSMUST00000066819.4 |
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chr17_-_26199008 | 1.04 |
ENSMUST00000142410.1
ENSMUST00000120333.1 ENSMUST00000039113.7 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr7_-_144939823 | 0.99 |
ENSMUST00000093962.4
|
Ccnd1
|
cyclin D1 |
| chr5_-_45639501 | 0.98 |
ENSMUST00000016023.7
|
Fam184b
|
family with sequence similarity 184, member B |
| chr6_-_38299236 | 0.98 |
ENSMUST00000058524.2
|
Zc3hav1l
|
zinc finger CCCH-type, antiviral 1-like |
| chr13_-_62858364 | 0.96 |
ENSMUST00000021907.7
|
Fbp2
|
fructose bisphosphatase 2 |
| chr9_+_107906866 | 0.95 |
ENSMUST00000035203.7
|
Mst1r
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr9_+_21368014 | 0.95 |
ENSMUST00000067646.4
ENSMUST00000115414.1 |
Ilf3
|
interleukin enhancer binding factor 3 |
| chr14_-_78536854 | 0.95 |
ENSMUST00000022593.5
|
Akap11
|
A kinase (PRKA) anchor protein 11 |
| chr1_+_59684949 | 0.93 |
ENSMUST00000027174.3
|
Nop58
|
NOP58 ribonucleoprotein |
| chrX_+_74329058 | 0.91 |
ENSMUST00000004326.3
|
Plxna3
|
plexin A3 |
| chr2_+_4300462 | 0.90 |
ENSMUST00000175669.1
|
Frmd4a
|
FERM domain containing 4A |
| chr9_-_35558522 | 0.89 |
ENSMUST00000034612.5
|
Ddx25
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 25 |
| chr6_+_17463826 | 0.87 |
ENSMUST00000140070.1
|
Met
|
met proto-oncogene |
| chrX_+_166344692 | 0.87 |
ENSMUST00000112223.1
ENSMUST00000112224.1 ENSMUST00000112229.2 ENSMUST00000112228.1 ENSMUST00000112227.2 ENSMUST00000112226.2 |
Gpm6b
|
glycoprotein m6b |
| chr4_+_11191354 | 0.86 |
ENSMUST00000170901.1
|
Ccne2
|
cyclin E2 |
| chr16_+_94370618 | 0.85 |
ENSMUST00000117648.1
ENSMUST00000147352.1 ENSMUST00000150346.1 ENSMUST00000155692.1 ENSMUST00000153988.1 ENSMUST00000141856.1 ENSMUST00000152117.1 ENSMUST00000150097.1 ENSMUST00000122895.1 ENSMUST00000151770.1 |
Ttc3
|
tetratricopeptide repeat domain 3 |
| chr14_-_78536762 | 0.84 |
ENSMUST00000123853.1
|
Akap11
|
A kinase (PRKA) anchor protein 11 |
| chr7_-_46795661 | 0.81 |
ENSMUST00000123725.1
|
Hps5
|
Hermansky-Pudlak syndrome 5 homolog (human) |
| chr3_+_159495408 | 0.81 |
ENSMUST00000120272.1
ENSMUST00000029825.7 ENSMUST00000106041.2 |
Depdc1a
|
DEP domain containing 1a |
| chr6_+_17463927 | 0.81 |
ENSMUST00000115442.1
|
Met
|
met proto-oncogene |
| chr5_+_67607873 | 0.79 |
ENSMUST00000087241.5
|
Shisa3
|
shisa homolog 3 (Xenopus laevis) |
| chr4_-_155992604 | 0.77 |
ENSMUST00000052185.3
|
B3galt6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase, polypeptide 6 |
| chr16_-_44139630 | 0.77 |
ENSMUST00000137557.1
ENSMUST00000147025.1 |
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr12_+_85746539 | 0.77 |
ENSMUST00000040461.3
|
Mfsd7c
|
major facilitator superfamily domain containing 7C |
| chr16_-_44139003 | 0.76 |
ENSMUST00000124102.1
|
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr10_+_13090788 | 0.75 |
ENSMUST00000121646.1
ENSMUST00000121325.1 ENSMUST00000121766.1 |
Plagl1
|
pleiomorphic adenoma gene-like 1 |
| chr7_+_47050628 | 0.75 |
ENSMUST00000010451.5
|
Tmem86a
|
transmembrane protein 86A |
| chr3_-_56183678 | 0.75 |
ENSMUST00000029374.6
|
Nbea
|
neurobeachin |
| chrX_+_153832225 | 0.74 |
ENSMUST00000148708.1
ENSMUST00000123264.1 ENSMUST00000049999.8 |
Spin2c
|
spindlin family, member 2C |
| chrX_+_36328353 | 0.71 |
ENSMUST00000016383.3
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr10_+_128909866 | 0.71 |
ENSMUST00000026407.7
|
Cd63
|
CD63 antigen |
| chr4_+_57568144 | 0.70 |
ENSMUST00000102904.3
|
Palm2
|
paralemmin 2 |
| chr16_+_94370786 | 0.69 |
ENSMUST00000147046.1
ENSMUST00000149885.1 ENSMUST00000127667.1 ENSMUST00000119131.1 ENSMUST00000145883.1 |
Ttc3
|
tetratricopeptide repeat domain 3 |
| chrX_-_85776606 | 0.69 |
ENSMUST00000142152.1
ENSMUST00000156390.1 ENSMUST00000113978.2 |
Gyk
|
glycerol kinase |
| chr6_+_4902913 | 0.68 |
ENSMUST00000175889.1
ENSMUST00000168998.2 |
Ppp1r9a
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A |
| chr4_+_148039097 | 0.67 |
ENSMUST00000141283.1
|
Mthfr
|
5,10-methylenetetrahydrofolate reductase |
| chr7_+_110122299 | 0.67 |
ENSMUST00000033326.8
|
Wee1
|
WEE 1 homolog 1 (S. pombe) |
| chr3_+_41564880 | 0.67 |
ENSMUST00000168086.1
|
Phf17
|
PHD finger protein 17 |
| chr7_-_4445181 | 0.66 |
ENSMUST00000138798.1
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chrX_-_151017251 | 0.66 |
ENSMUST00000112691.2
ENSMUST00000026297.5 ENSMUST00000154393.1 ENSMUST00000156233.1 |
Gnl3l
|
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr2_-_37703845 | 0.66 |
ENSMUST00000155237.1
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr10_+_67535465 | 0.66 |
ENSMUST00000145754.1
|
Egr2
|
early growth response 2 |
| chr3_+_152396664 | 0.65 |
ENSMUST00000089982.4
ENSMUST00000106101.1 |
Zzz3
|
zinc finger, ZZ domain containing 3 |
| chr11_-_102819663 | 0.65 |
ENSMUST00000092567.4
|
Gjc1
|
gap junction protein, gamma 1 |
| chr10_+_36974558 | 0.65 |
ENSMUST00000105510.1
|
Hdac2
|
histone deacetylase 2 |
| chr10_-_69212996 | 0.65 |
ENSMUST00000170048.1
|
A930033H14Rik
|
RIKEN cDNA A930033H14 gene |
| chr15_+_44787746 | 0.65 |
ENSMUST00000181839.1
|
2310069G16Rik
|
RIKEN cDNA 2310069G16 gene |
| chr13_+_73626886 | 0.65 |
ENSMUST00000022104.7
|
Tert
|
telomerase reverse transcriptase |
| chr12_+_102128718 | 0.64 |
ENSMUST00000159329.1
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr10_-_4432285 | 0.63 |
ENSMUST00000155172.1
|
Rmnd1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae) |
| chr7_+_66839726 | 0.63 |
ENSMUST00000098382.3
|
Adamts17
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 17 |
| chr3_+_135826075 | 0.63 |
ENSMUST00000029810.5
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr5_-_148995147 | 0.63 |
ENSMUST00000147473.1
|
Katnal1
|
katanin p60 subunit A-like 1 |
| chr18_+_65581704 | 0.63 |
ENSMUST00000182979.1
|
Zfp532
|
zinc finger protein 532 |
| chr17_+_87107621 | 0.62 |
ENSMUST00000041369.6
|
Socs5
|
suppressor of cytokine signaling 5 |
| chr5_+_99979061 | 0.61 |
ENSMUST00000046721.1
|
4930524J08Rik
|
RIKEN cDNA 4930524J08 gene |
| chr10_+_95417352 | 0.61 |
ENSMUST00000181781.1
|
5730420D15Rik
|
RIKEN cDNA 5730420D15 gene |
| chr3_+_88532314 | 0.61 |
ENSMUST00000172699.1
|
Mex3a
|
mex3 homolog A (C. elegans) |
| chr16_+_31663841 | 0.59 |
ENSMUST00000115201.1
|
Dlg1
|
discs, large homolog 1 (Drosophila) |
| chrX_-_93632113 | 0.58 |
ENSMUST00000006856.2
|
Pola1
|
polymerase (DNA directed), alpha 1 |
| chr4_-_133756769 | 0.58 |
ENSMUST00000008024.6
|
Arid1a
|
AT rich interactive domain 1A (SWI-like) |
| chr2_+_30286383 | 0.57 |
ENSMUST00000064447.5
|
Nup188
|
nucleoporin 188 |
| chr4_+_131921771 | 0.57 |
ENSMUST00000094666.3
|
Tmem200b
|
transmembrane protein 200B |
| chr3_+_135825648 | 0.57 |
ENSMUST00000180196.1
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr3_-_75956888 | 0.56 |
ENSMUST00000038563.7
ENSMUST00000167078.1 ENSMUST00000117242.1 |
Golim4
|
golgi integral membrane protein 4 |
| chrX_-_51681703 | 0.56 |
ENSMUST00000088172.5
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chrX_+_136270302 | 0.56 |
ENSMUST00000113112.1
|
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr19_+_10018265 | 0.55 |
ENSMUST00000131407.1
|
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr5_+_105415738 | 0.54 |
ENSMUST00000112707.1
|
Lrrc8b
|
leucine rich repeat containing 8 family, member B |
| chr10_-_127180579 | 0.54 |
ENSMUST00000095270.2
|
Slc26a10
|
solute carrier family 26, member 10 |
| chr11_+_69935796 | 0.54 |
ENSMUST00000018698.5
|
Ybx2
|
Y box protein 2 |
| chrX_+_81070646 | 0.54 |
ENSMUST00000171953.1
ENSMUST00000026760.2 |
Tmem47
|
transmembrane protein 47 |
| chr8_+_88272403 | 0.54 |
ENSMUST00000169037.1
|
Adcy7
|
adenylate cyclase 7 |
| chr1_-_128592284 | 0.54 |
ENSMUST00000052172.6
ENSMUST00000142893.1 |
Cxcr4
|
chemokine (C-X-C motif) receptor 4 |
| chr5_+_76951382 | 0.53 |
ENSMUST00000141687.1
|
Paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase |
| chr3_+_28781305 | 0.53 |
ENSMUST00000060500.7
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr1_+_87327008 | 0.53 |
ENSMUST00000172794.1
ENSMUST00000164992.2 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr15_-_10714612 | 0.53 |
ENSMUST00000169385.1
|
Rai14
|
retinoic acid induced 14 |
| chr3_+_122274371 | 0.53 |
ENSMUST00000035776.8
|
Dnttip2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr2_-_92370999 | 0.53 |
ENSMUST00000176810.1
ENSMUST00000090582.4 |
Gyltl1b
|
glycosyltransferase-like 1B |
| chr19_-_10203880 | 0.53 |
ENSMUST00000142241.1
ENSMUST00000116542.2 ENSMUST00000025651.5 ENSMUST00000156291.1 |
Fen1
|
flap structure specific endonuclease 1 |
| chr4_-_53159885 | 0.53 |
ENSMUST00000030010.3
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr9_-_121495678 | 0.52 |
ENSMUST00000035120.4
|
Cck
|
cholecystokinin |
| chrX_-_162829379 | 0.52 |
ENSMUST00000041370.4
ENSMUST00000112316.2 ENSMUST00000112315.1 |
Txlng
|
taxilin gamma |
| chr8_+_70234613 | 0.52 |
ENSMUST00000145078.1
|
Sugp2
|
SURP and G patch domain containing 2 |
| chr2_+_48949495 | 0.52 |
ENSMUST00000112745.1
|
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr10_-_19015347 | 0.52 |
ENSMUST00000019997.4
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr10_+_4266323 | 0.52 |
ENSMUST00000045730.5
|
Akap12
|
A kinase (PRKA) anchor protein (gravin) 12 |
| chr2_+_160645881 | 0.52 |
ENSMUST00000109468.2
|
Top1
|
topoisomerase (DNA) I |
| chr11_-_106789157 | 0.51 |
ENSMUST00000129585.1
|
Ddx5
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 |
| chr7_-_82648469 | 0.51 |
ENSMUST00000056728.4
|
Fam154b
|
family with sequence similarity 154, member B |
| chr11_-_106788845 | 0.51 |
ENSMUST00000123339.1
|
Ddx5
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 |
| chrX_+_166238923 | 0.51 |
ENSMUST00000060210.7
ENSMUST00000112233.1 |
Gpm6b
|
glycoprotein m6b |
| chr11_+_76217608 | 0.51 |
ENSMUST00000040806.4
|
Dbil5
|
diazepam binding inhibitor-like 5 |
| chr1_+_87327044 | 0.50 |
ENSMUST00000173173.1
|
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr19_-_16780822 | 0.50 |
ENSMUST00000068156.6
|
Vps13a
|
vacuolar protein sorting 13A (yeast) |
| chr3_+_135825788 | 0.50 |
ENSMUST00000167390.1
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr19_-_4615647 | 0.50 |
ENSMUST00000113822.2
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr7_+_98703091 | 0.50 |
ENSMUST00000033009.9
|
Prkrir
|
protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor) |
| chr13_-_119408985 | 0.49 |
ENSMUST00000099149.3
ENSMUST00000069902.6 ENSMUST00000109204.1 |
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr7_+_66839752 | 0.49 |
ENSMUST00000107478.1
|
Adamts17
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 17 |
| chr4_+_130055010 | 0.49 |
ENSMUST00000123617.1
|
Col16a1
|
collagen, type XVI, alpha 1 |
| chr2_+_181493185 | 0.49 |
ENSMUST00000069649.8
|
Abhd16b
|
abhydrolase domain containing 16B |
| chr4_-_130574150 | 0.48 |
ENSMUST00000105993.3
|
Nkain1
|
Na+/K+ transporting ATPase interacting 1 |
| chr11_+_44518959 | 0.48 |
ENSMUST00000019333.3
|
Rnf145
|
ring finger protein 145 |
| chr4_+_44756609 | 0.48 |
ENSMUST00000143385.1
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr14_+_31019183 | 0.48 |
ENSMUST00000052239.5
|
Pbrm1
|
polybromo 1 |
| chr19_-_4615453 | 0.48 |
ENSMUST00000053597.2
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr7_-_4445595 | 0.48 |
ENSMUST00000119485.1
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr6_+_5390387 | 0.48 |
ENSMUST00000183358.1
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr1_-_186705980 | 0.47 |
ENSMUST00000045288.8
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr9_+_44066993 | 0.47 |
ENSMUST00000034508.7
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr4_+_74251928 | 0.47 |
ENSMUST00000030102.5
|
Kdm4c
|
lysine (K)-specific demethylase 4C |
| chr12_-_102878406 | 0.47 |
ENSMUST00000045652.6
|
Btbd7
|
BTB (POZ) domain containing 7 |
| chr8_-_122551316 | 0.46 |
ENSMUST00000067252.7
|
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr4_+_32657107 | 0.46 |
ENSMUST00000071642.4
ENSMUST00000178134.1 |
Mdn1
|
midasin homolog (yeast) |
| chr13_-_55329723 | 0.46 |
ENSMUST00000021941.7
|
Mxd3
|
Max dimerization protein 3 |
| chr2_+_124089961 | 0.46 |
ENSMUST00000103241.1
|
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr13_+_37826225 | 0.46 |
ENSMUST00000128570.1
|
Rreb1
|
ras responsive element binding protein 1 |
| chr19_-_29805507 | 0.45 |
ENSMUST00000175726.1
|
9930021J03Rik
|
RIKEN cDNA 9930021J03 gene |
| chr9_+_44067072 | 0.45 |
ENSMUST00000177054.1
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr5_+_100039990 | 0.45 |
ENSMUST00000169390.1
ENSMUST00000031268.6 |
Enoph1
|
enolase-phosphatase 1 |
| chr5_-_72168142 | 0.45 |
ENSMUST00000013693.6
|
Commd8
|
COMM domain containing 8 |
| chr10_+_36974536 | 0.45 |
ENSMUST00000019911.7
|
Hdac2
|
histone deacetylase 2 |
| chr2_-_92371039 | 0.44 |
ENSMUST00000068586.6
|
Gyltl1b
|
glycosyltransferase-like 1B |
| chr14_+_31019159 | 0.44 |
ENSMUST00000112094.1
ENSMUST00000144009.1 |
Pbrm1
|
polybromo 1 |
| chrX_-_7572843 | 0.44 |
ENSMUST00000132788.1
|
Ppp1r3f
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F |
| chr1_-_66817536 | 0.44 |
ENSMUST00000068168.3
ENSMUST00000113987.1 |
Kansl1l
|
KAT8 regulatory NSL complex subunit 1-like |
| chr15_-_53902472 | 0.44 |
ENSMUST00000078673.6
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr11_+_69935894 | 0.44 |
ENSMUST00000149194.1
|
Ybx2
|
Y box protein 2 |
| chr1_-_152766323 | 0.44 |
ENSMUST00000111857.1
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr1_+_183297060 | 0.44 |
ENSMUST00000109166.2
|
Aida
|
axin interactor, dorsalization associated |
| chr10_-_80139347 | 0.44 |
ENSMUST00000105369.1
|
Dos
|
downstream of Stk11 |
| chr7_-_133123770 | 0.43 |
ENSMUST00000164896.1
ENSMUST00000171968.1 |
Ctbp2
|
C-terminal binding protein 2 |
| chr17_-_6477102 | 0.43 |
ENSMUST00000167717.2
|
Tmem181b-ps
|
transmembrane protein 181B, pseudogene |
| chr2_+_83812567 | 0.43 |
ENSMUST00000051454.3
|
Fam171b
|
family with sequence similarity 171, member B |
| chr4_+_148039035 | 0.42 |
ENSMUST00000097788.4
|
Mthfr
|
5,10-methylenetetrahydrofolate reductase |
| chr2_+_130274437 | 0.42 |
ENSMUST00000141872.1
|
Nop56
|
NOP56 ribonucleoprotein |
| chr18_-_12941777 | 0.42 |
ENSMUST00000122175.1
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr7_-_4445637 | 0.42 |
ENSMUST00000008579.7
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr13_-_48625571 | 0.42 |
ENSMUST00000035824.9
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr15_+_102296256 | 0.41 |
ENSMUST00000064924.4
|
Espl1
|
extra spindle poles-like 1 (S. cerevisiae) |
| chr9_-_123215942 | 0.41 |
ENSMUST00000039229.7
|
Cdcp1
|
CUB domain containing protein 1 |
| chr5_+_143933059 | 0.41 |
ENSMUST00000166847.1
|
Rsph10b
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chrX_+_134686519 | 0.40 |
ENSMUST00000124226.2
|
Armcx4
|
armadillo repeat containing, X-linked 4 |
| chr11_+_69098937 | 0.40 |
ENSMUST00000021271.7
|
Per1
|
period circadian clock 1 |
| chr11_+_49203465 | 0.40 |
ENSMUST00000150284.1
ENSMUST00000109197.1 ENSMUST00000151228.1 |
Zfp62
|
zinc finger protein 62 |
| chr14_-_21052452 | 0.40 |
ENSMUST00000130291.1
|
Ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr10_+_67535493 | 0.40 |
ENSMUST00000145936.1
|
Egr2
|
early growth response 2 |
| chr2_-_92370968 | 0.40 |
ENSMUST00000176774.1
|
Gyltl1b
|
glycosyltransferase-like 1B |
| chr6_+_4903298 | 0.40 |
ENSMUST00000035813.2
|
Ppp1r9a
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A |
| chr11_-_86757483 | 0.40 |
ENSMUST00000060766.9
ENSMUST00000103186.4 |
Cltc
|
clathrin, heavy polypeptide (Hc) |
| chr11_-_102819114 | 0.40 |
ENSMUST00000068933.5
|
Gjc1
|
gap junction protein, gamma 1 |
| chr16_-_43889669 | 0.40 |
ENSMUST00000023387.7
|
Qtrtd1
|
queuine tRNA-ribosyltransferase domain containing 1 |
| chr13_-_9764865 | 0.39 |
ENSMUST00000128658.1
|
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr12_+_71136848 | 0.39 |
ENSMUST00000149564.1
ENSMUST00000045907.8 |
2700049A03Rik
|
RIKEN cDNA 2700049A03 gene |
| chr17_+_25133385 | 0.39 |
ENSMUST00000040729.2
|
Clcn7
|
chloride channel 7 |
| chr2_+_132816141 | 0.39 |
ENSMUST00000028831.8
ENSMUST00000066559.5 |
Mcm8
|
minichromosome maintenance deficient 8 (S. cerevisiae) |
| chr15_-_79834261 | 0.39 |
ENSMUST00000148358.1
|
Cbx6
|
chromobox 6 |
| chr11_+_60353324 | 0.39 |
ENSMUST00000070805.6
ENSMUST00000094140.2 ENSMUST00000108723.2 ENSMUST00000108722.4 |
Lrrc48
|
leucine rich repeat containing 48 |
| chr19_-_60581013 | 0.39 |
ENSMUST00000111460.3
ENSMUST00000081790.7 ENSMUST00000166712.1 |
Cacul1
|
CDK2 associated, cullin domain 1 |
| chr16_-_45158183 | 0.39 |
ENSMUST00000114600.1
|
Slc35a5
|
solute carrier family 35, member A5 |
| chrX_+_163909132 | 0.39 |
ENSMUST00000033734.7
ENSMUST00000112294.2 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr5_-_100500592 | 0.38 |
ENSMUST00000149714.1
ENSMUST00000046154.5 |
Lin54
|
lin-54 homolog (C. elegans) |
| chr4_+_134468320 | 0.38 |
ENSMUST00000030636.4
ENSMUST00000127279.1 ENSMUST00000105867.1 |
Stmn1
|
stathmin 1 |
| chr19_-_58860975 | 0.38 |
ENSMUST00000066285.4
|
Hspa12a
|
heat shock protein 12A |
| chr1_+_93135244 | 0.38 |
ENSMUST00000027491.5
|
Agxt
|
alanine-glyoxylate aminotransferase |
| chr13_+_5861489 | 0.38 |
ENSMUST00000000080.6
|
Klf6
|
Kruppel-like factor 6 |
| chr7_-_46795881 | 0.38 |
ENSMUST00000107653.1
ENSMUST00000107654.1 ENSMUST00000014562.7 ENSMUST00000152759.1 |
Hps5
|
Hermansky-Pudlak syndrome 5 homolog (human) |
| chr2_+_156196642 | 0.38 |
ENSMUST00000037401.8
|
Phf20
|
PHD finger protein 20 |
| chr1_-_153549697 | 0.37 |
ENSMUST00000041874.7
|
Npl
|
N-acetylneuraminate pyruvate lyase |
| chr1_-_55088156 | 0.37 |
ENSMUST00000127861.1
ENSMUST00000144077.1 |
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr11_-_115027012 | 0.37 |
ENSMUST00000100240.2
|
Gm11710
|
predicted gene 11710 |
| chr19_-_60790692 | 0.37 |
ENSMUST00000025955.6
|
Eif3a
|
eukaryotic translation initiation factor 3, subunit A |
| chr17_+_56040350 | 0.37 |
ENSMUST00000002914.8
|
Chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr10_-_78464853 | 0.37 |
ENSMUST00000105385.1
|
Pdxk
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr6_-_88898664 | 0.37 |
ENSMUST00000058011.6
|
Mcm2
|
minichromosome maintenance deficient 2 mitotin (S. cerevisiae) |
| chr11_-_84513485 | 0.37 |
ENSMUST00000018841.2
|
Aatf
|
apoptosis antagonizing transcription factor |
| chrX_-_12160355 | 0.37 |
ENSMUST00000043441.6
|
Bcor
|
BCL6 interacting corepressor |
| chr18_+_53176345 | 0.37 |
ENSMUST00000037850.5
|
Snx2
|
sorting nexin 2 |
| chr14_-_36935560 | 0.37 |
ENSMUST00000183038.1
|
Ccser2
|
coiled-coil serine rich 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mnt
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1904170 | regulation of bleb assembly(GO:1904170) |
| 0.3 | 1.7 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.3 | 2.2 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.3 | 1.2 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.3 | 1.5 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) |
| 0.3 | 1.7 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 | 1.1 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.3 | 1.1 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.3 | 1.1 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.3 | 1.1 | GO:0021594 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.3 | 1.0 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.3 | 0.8 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 0.2 | 1.0 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.2 | 1.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 0.7 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.2 | 1.1 | GO:0035983 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.2 | 0.6 | GO:1904173 | regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 0.2 | 0.5 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.2 | 0.5 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 1.6 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.2 | 0.5 | GO:0002632 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.2 | 0.7 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.2 | 0.5 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.2 | 1.5 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.2 | 2.2 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.2 | 1.6 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.2 | 0.5 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.2 | 0.9 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.2 | 0.9 | GO:1902287 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.6 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.1 | 0.6 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.1 | 0.4 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.1 | 0.6 | GO:0006272 | DNA replication, synthesis of RNA primer(GO:0006269) leading strand elongation(GO:0006272) |
| 0.1 | 0.7 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 1.0 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 | 0.5 | GO:0060155 | response to high density lipoprotein particle(GO:0055099) platelet dense granule organization(GO:0060155) |
| 0.1 | 0.4 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 0.6 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.3 | GO:0021571 | rhombomere 3 development(GO:0021569) rhombomere 5 development(GO:0021571) |
| 0.1 | 0.3 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.1 | 0.6 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.4 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.3 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.1 | 0.7 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.1 | 0.2 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.3 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.6 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 1.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 0.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.6 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.4 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.5 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.9 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.2 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 0.3 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.6 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 1.8 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 1.9 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 0.2 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.1 | 0.3 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.1 | 0.4 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.2 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.2 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.1 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 0.2 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.3 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 0.1 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.5 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.6 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.1 | 0.3 | GO:0046073 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 0.1 | 0.6 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.1 | 0.3 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.1 | 0.2 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.1 | 0.7 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.6 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.1 | GO:0003284 | septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) |
| 0.1 | 0.5 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.3 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 0.2 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.1 | 0.5 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.1 | 1.3 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.3 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.4 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.1 | 0.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.3 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.3 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.1 | 0.5 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 1.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.5 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 1.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.5 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.3 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.2 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.8 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.2 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.0 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.2 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.2 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.2 | GO:0061295 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.0 | 0.5 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.4 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 0.3 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.5 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 | 0.8 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0061146 | midgut development(GO:0007494) Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.4 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.2 | GO:1901909 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.2 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.0 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 1.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.3 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 1.1 | GO:0043489 | RNA stabilization(GO:0043489) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0014858 | positive regulation of skeletal muscle cell proliferation(GO:0014858) |
| 0.0 | 0.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.5 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.9 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 1.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.4 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.1 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.0 | 0.6 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.5 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.3 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.5 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.5 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.2 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.5 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.1 | GO:0061738 | late endosomal microautophagy(GO:0061738) protein targeting to vacuole involved in autophagy(GO:0071211) lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.0 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.0 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.2 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.2 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.3 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.4 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.0 | 0.5 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.2 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 1.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 1.1 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0090177 | establishment of planar polarity involved in neural tube closure(GO:0090177) |
| 0.0 | 0.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 1.4 | GO:0010771 | negative regulation of cell morphogenesis involved in differentiation(GO:0010771) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.0 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.5 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.3 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.0 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.3 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.1 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 1.2 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.3 | 1.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.3 | 0.9 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.3 | 0.8 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.2 | 1.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.2 | 1.0 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 1.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 0.7 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.2 | 1.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 1.8 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.6 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 1.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.7 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.4 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 0.7 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 1.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 1.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.5 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 0.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.3 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 0.4 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 0.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.4 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 1.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.1 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.1 | 0.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 2.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.6 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.5 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 2.0 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 1.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.9 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.7 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0000802 | transverse filament(GO:0000802) |
| 0.0 | 0.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.2 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 1.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.0 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.0 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 1.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.0 | 1.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 1.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.5 | 1.8 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.4 | 1.2 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.4 | 1.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.3 | 1.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 1.1 | GO:0035851 | histone deacetylase activity (H4-K16 specific)(GO:0034739) Krueppel-associated box domain binding(GO:0035851) |
| 0.3 | 1.0 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.3 | 0.8 | GO:0017084 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 0.2 | 1.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.2 | 0.6 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.2 | 1.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 0.5 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.1 | 0.6 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.6 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 1.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.5 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.1 | 0.8 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.6 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.1 | 0.4 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.1 | 0.9 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.5 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.9 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.9 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 0.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.3 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.1 | 2.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.4 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 2.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.8 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.3 | GO:0070905 | serine binding(GO:0070905) |
| 0.1 | 0.3 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 1.1 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.5 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.6 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 1.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 0.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.4 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.1 | 1.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.8 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 0.2 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.1 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.4 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.2 | GO:0016822 | oxaloacetate decarboxylase activity(GO:0008948) hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.5 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 2.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 1.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 2.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.3 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.2 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.4 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 1.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.5 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.0 | 0.8 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.2 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.3 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 1.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.0 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 1.1 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0038100 | nodal binding(GO:0038100) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.2 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.0 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 0.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 2.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.2 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.1 | 0.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 1.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.9 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 0.8 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 3.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.8 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 2.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME DNA STRAND ELONGATION | Genes involved in DNA strand elongation |
| 0.0 | 1.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 2.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.7 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.2 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.5 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |