Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
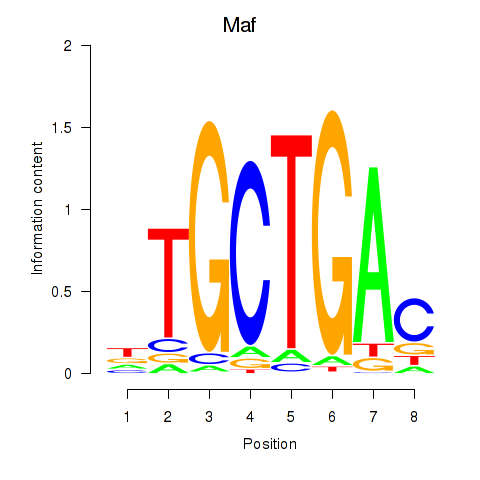
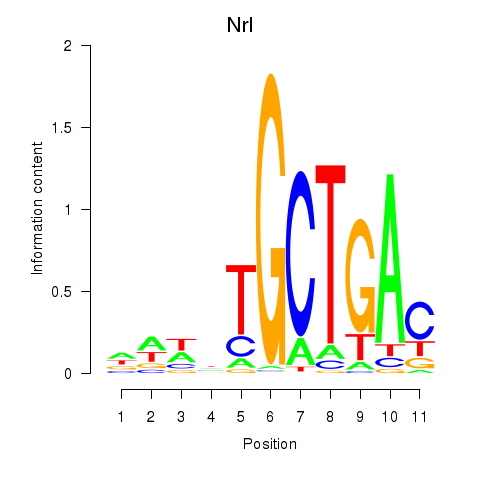
Results for Maf_Nrl
Z-value: 1.64
Transcription factors associated with Maf_Nrl
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Maf
|
ENSMUSG00000055435.6 | avian musculoaponeurotic fibrosarcoma oncogene homolog |
|
Nrl
|
ENSMUSG00000040632.9 | neural retina leucine zipper gene |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nrl | mm10_v2_chr14_-_55524938_55524967 | -0.38 | 2.2e-02 | Click! |
| Maf | mm10_v2_chr8_-_115706994_115707096 | -0.38 | 2.4e-02 | Click! |
Activity profile of Maf_Nrl motif
Sorted Z-values of Maf_Nrl motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_39007019 | 13.61 |
ENSMUST00000025966.4
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr10_+_87860030 | 13.41 |
ENSMUST00000062862.6
|
Igf1
|
insulin-like growth factor 1 |
| chr10_+_87859593 | 12.31 |
ENSMUST00000126490.1
|
Igf1
|
insulin-like growth factor 1 |
| chr10_+_87859481 | 7.64 |
ENSMUST00000121952.1
|
Igf1
|
insulin-like growth factor 1 |
| chr10_+_87859255 | 6.87 |
ENSMUST00000105300.2
|
Igf1
|
insulin-like growth factor 1 |
| chr10_+_87859062 | 6.48 |
ENSMUST00000095360.4
|
Igf1
|
insulin-like growth factor 1 |
| chr9_-_46235260 | 6.29 |
ENSMUST00000121916.1
ENSMUST00000034586.2 |
Apoc3
|
apolipoprotein C-III |
| chr19_+_20601958 | 5.65 |
ENSMUST00000087638.3
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr19_-_20727533 | 5.17 |
ENSMUST00000025656.3
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr4_+_104766334 | 4.55 |
ENSMUST00000065072.6
|
C8b
|
complement component 8, beta polypeptide |
| chr4_+_104766308 | 4.49 |
ENSMUST00000031663.3
|
C8b
|
complement component 8, beta polypeptide |
| chr8_-_3878549 | 3.59 |
ENSMUST00000011445.6
|
Cd209d
|
CD209d antigen |
| chr4_+_141239499 | 3.41 |
ENSMUST00000141834.2
|
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chrX_+_42502533 | 3.34 |
ENSMUST00000005839.4
|
Sh2d1a
|
SH2 domain protein 1A |
| chr19_-_21652779 | 3.19 |
ENSMUST00000179768.1
ENSMUST00000178523.1 ENSMUST00000038830.3 |
1110059E24Rik
|
RIKEN cDNA 1110059E24 gene |
| chr9_-_46235631 | 3.13 |
ENSMUST00000118649.1
|
Apoc3
|
apolipoprotein C-III |
| chr1_-_136260873 | 3.09 |
ENSMUST00000086395.5
|
Gpr25
|
G protein-coupled receptor 25 |
| chr4_-_82705735 | 2.61 |
ENSMUST00000155821.1
|
Nfib
|
nuclear factor I/B |
| chr14_+_37068042 | 2.55 |
ENSMUST00000057176.3
|
Lrit2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 |
| chr7_+_51878967 | 2.54 |
ENSMUST00000051912.6
|
Gas2
|
growth arrest specific 2 |
| chr9_-_70141484 | 2.48 |
ENSMUST00000034749.8
|
Fam81a
|
family with sequence similarity 81, member A |
| chr15_+_10223974 | 2.48 |
ENSMUST00000128450.1
ENSMUST00000148257.1 ENSMUST00000128921.1 |
Prlr
|
prolactin receptor |
| chr16_+_20733104 | 2.41 |
ENSMUST00000115423.1
ENSMUST00000007171.6 |
Chrd
|
chordin |
| chr7_+_51879041 | 2.35 |
ENSMUST00000107591.2
|
Gas2
|
growth arrest specific 2 |
| chr5_-_113081579 | 2.31 |
ENSMUST00000131708.1
ENSMUST00000117143.1 ENSMUST00000119627.1 |
Crybb3
|
crystallin, beta B3 |
| chr7_-_4789541 | 2.13 |
ENSMUST00000168578.1
|
Tmem238
|
transmembrane protein 238 |
| chr14_-_37048957 | 2.10 |
ENSMUST00000022338.5
|
Rgr
|
retinal G protein coupled receptor |
| chr17_+_12584183 | 2.03 |
ENSMUST00000046959.7
|
Slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr8_-_3926798 | 2.02 |
ENSMUST00000171635.1
ENSMUST00000111014.1 ENSMUST00000084086.2 |
Cd209b
|
CD209b antigen |
| chr1_+_74284930 | 1.96 |
ENSMUST00000113805.1
ENSMUST00000027370.6 ENSMUST00000087226.4 |
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr14_-_73385225 | 1.91 |
ENSMUST00000022704.7
|
Itm2b
|
integral membrane protein 2B |
| chr4_+_141242850 | 1.89 |
ENSMUST00000138096.1
ENSMUST00000006618.2 ENSMUST00000125392.1 |
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr16_-_46155077 | 1.87 |
ENSMUST00000059524.5
|
Gm4737
|
predicted gene 4737 |
| chrX_+_42502596 | 1.84 |
ENSMUST00000115070.1
ENSMUST00000153948.1 |
Sh2d1a
|
SH2 domain protein 1A |
| chr10_+_4710119 | 1.83 |
ENSMUST00000105588.1
ENSMUST00000105589.1 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr17_+_46428930 | 1.82 |
ENSMUST00000024764.5
ENSMUST00000165993.1 |
Crip3
|
cysteine-rich protein 3 |
| chr4_-_141239453 | 1.79 |
ENSMUST00000168138.1
|
C630004L07Rik
|
RIKEN cDNA C630004L07 gene |
| chr17_+_46428917 | 1.72 |
ENSMUST00000113465.3
|
Crip3
|
cysteine-rich protein 3 |
| chr1_+_127729405 | 1.71 |
ENSMUST00000038006.6
|
Acmsd
|
amino carboxymuconate semialdehyde decarboxylase |
| chr4_+_143349757 | 1.61 |
ENSMUST00000052458.2
|
Lrrc38
|
leucine rich repeat containing 38 |
| chr1_+_88095054 | 1.58 |
ENSMUST00000150634.1
ENSMUST00000058237.7 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr13_-_64129305 | 1.58 |
ENSMUST00000099441.4
|
Slc35d2
|
solute carrier family 35, member D2 |
| chr16_+_64851991 | 1.58 |
ENSMUST00000067744.7
|
Cggbp1
|
CGG triplet repeat binding protein 1 |
| chr3_+_135826075 | 1.55 |
ENSMUST00000029810.5
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr6_-_29380426 | 1.53 |
ENSMUST00000147483.2
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr11_-_53423123 | 1.51 |
ENSMUST00000036045.5
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr11_-_77725281 | 1.51 |
ENSMUST00000078623.4
|
Cryba1
|
crystallin, beta A1 |
| chr8_+_119437118 | 1.50 |
ENSMUST00000152420.1
ENSMUST00000098365.3 |
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr6_+_17491216 | 1.45 |
ENSMUST00000080469.5
|
Met
|
met proto-oncogene |
| chr2_+_69897220 | 1.43 |
ENSMUST00000055758.9
ENSMUST00000112251.2 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr13_+_48261427 | 1.41 |
ENSMUST00000021810.1
|
Id4
|
inhibitor of DNA binding 4 |
| chr9_-_107679592 | 1.38 |
ENSMUST00000010205.7
|
Gnat1
|
guanine nucleotide binding protein, alpha transducing 1 |
| chr5_-_34187670 | 1.38 |
ENSMUST00000042701.6
ENSMUST00000119171.1 |
Mxd4
|
Max dimerization protein 4 |
| chr10_-_24101951 | 1.37 |
ENSMUST00000170267.1
|
Taar8c
|
trace amine-associated receptor 8C |
| chrX_-_143933204 | 1.37 |
ENSMUST00000112851.1
ENSMUST00000112856.2 ENSMUST00000033642.3 |
Dcx
|
doublecortin |
| chr6_+_113472276 | 1.32 |
ENSMUST00000147316.1
|
Il17rc
|
interleukin 17 receptor C |
| chr19_+_37685581 | 1.28 |
ENSMUST00000073391.4
|
Cyp26c1
|
cytochrome P450, family 26, subfamily c, polypeptide 1 |
| chr3_+_94398517 | 1.27 |
ENSMUST00000050975.3
|
Lingo4
|
leucine rich repeat and Ig domain containing 4 |
| chr2_-_148040196 | 1.27 |
ENSMUST00000136555.1
|
9030622O22Rik
|
RIKEN cDNA 9030622O22 gene |
| chr9_+_46228580 | 1.26 |
ENSMUST00000034588.8
|
Apoa1
|
apolipoprotein A-I |
| chr11_-_50210765 | 1.25 |
ENSMUST00000143379.1
ENSMUST00000015981.5 ENSMUST00000102774.4 |
Sqstm1
|
sequestosome 1 |
| chr4_-_6275629 | 1.21 |
ENSMUST00000029905.1
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr5_-_77408034 | 1.20 |
ENSMUST00000163898.1
ENSMUST00000046746.6 |
Igfbp7
|
insulin-like growth factor binding protein 7 |
| chr19_-_24861828 | 1.19 |
ENSMUST00000047666.4
|
Pgm5
|
phosphoglucomutase 5 |
| chr5_-_139813237 | 1.19 |
ENSMUST00000110832.1
|
Tmem184a
|
transmembrane protein 184a |
| chr6_+_71199827 | 1.16 |
ENSMUST00000067492.7
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr7_+_19228334 | 1.16 |
ENSMUST00000063976.8
|
Opa3
|
optic atrophy 3 |
| chr2_+_34772089 | 1.16 |
ENSMUST00000028222.6
ENSMUST00000100171.2 |
Hspa5
|
heat shock protein 5 |
| chr11_-_75422524 | 1.16 |
ENSMUST00000125982.1
ENSMUST00000137103.1 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr17_+_33920522 | 1.15 |
ENSMUST00000172489.1
|
Tapbp
|
TAP binding protein |
| chr15_+_57694651 | 1.15 |
ENSMUST00000096430.4
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr3_-_33083016 | 1.14 |
ENSMUST00000078226.3
ENSMUST00000108224.1 |
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr1_-_183369529 | 1.14 |
ENSMUST00000069922.5
|
Mia3
|
melanoma inhibitory activity 3 |
| chr6_+_117168535 | 1.12 |
ENSMUST00000112866.1
ENSMUST00000112871.1 ENSMUST00000073043.4 |
Cxcl12
|
chemokine (C-X-C motif) ligand 12 |
| chr7_+_101321703 | 1.10 |
ENSMUST00000174291.1
ENSMUST00000167888.2 ENSMUST00000172662.1 ENSMUST00000173270.1 ENSMUST00000174083.1 |
Stard10
|
START domain containing 10 |
| chr5_+_102845007 | 1.10 |
ENSMUST00000070000.4
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr6_+_90462562 | 1.10 |
ENSMUST00000032174.5
|
Klf15
|
Kruppel-like factor 15 |
| chr4_-_57300362 | 1.10 |
ENSMUST00000153926.1
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr2_+_101624696 | 1.10 |
ENSMUST00000044031.3
|
Rag2
|
recombination activating gene 2 |
| chr2_+_59160884 | 1.08 |
ENSMUST00000037903.8
|
Pkp4
|
plakophilin 4 |
| chr6_-_29380513 | 1.05 |
ENSMUST00000080428.6
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr14_+_54254124 | 1.04 |
ENSMUST00000180359.1
|
Abhd4
|
abhydrolase domain containing 4 |
| chr17_-_34031544 | 1.04 |
ENSMUST00000025186.8
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr1_+_59256906 | 1.03 |
ENSMUST00000160662.1
ENSMUST00000114248.2 |
Cdk15
|
cyclin-dependent kinase 15 |
| chr4_+_56740070 | 1.03 |
ENSMUST00000181745.1
|
Gm26657
|
predicted gene, 26657 |
| chr2_+_126034967 | 1.03 |
ENSMUST00000110442.1
|
Fgf7
|
fibroblast growth factor 7 |
| chr12_+_105336922 | 1.02 |
ENSMUST00000180503.1
|
2810011L19Rik
|
RIKEN cDNA 2810011L19 gene |
| chr3_-_97297778 | 1.02 |
ENSMUST00000181368.1
|
Gm17608
|
predicted gene, 17608 |
| chr7_+_121707189 | 1.01 |
ENSMUST00000065310.2
|
1700069B07Rik
|
RIKEN cDNA 1700069B07 gene |
| chr2_-_156887056 | 1.00 |
ENSMUST00000029164.2
|
Sla2
|
Src-like-adaptor 2 |
| chr17_-_34031684 | 1.00 |
ENSMUST00000169397.1
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr1_-_153851189 | 0.99 |
ENSMUST00000059607.6
|
5830403L16Rik
|
RIKEN cDNA 5830403L16 gene |
| chr17_-_34031644 | 0.99 |
ENSMUST00000171872.1
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr6_+_83054653 | 0.98 |
ENSMUST00000092618.6
|
Aup1
|
ancient ubiquitous protein 1 |
| chr3_-_84220853 | 0.96 |
ENSMUST00000154152.1
ENSMUST00000107693.2 ENSMUST00000107695.2 |
Trim2
|
tripartite motif-containing 2 |
| chr5_+_31116702 | 0.93 |
ENSMUST00000013771.8
|
Trim54
|
tripartite motif-containing 54 |
| chr17_-_26069409 | 0.92 |
ENSMUST00000120691.1
|
Rab11fip3
|
RAB11 family interacting protein 3 (class II) |
| chr11_+_69991633 | 0.92 |
ENSMUST00000108592.1
|
Gabarap
|
gamma-aminobutyric acid receptor associated protein |
| chr2_-_32694120 | 0.91 |
ENSMUST00000028148.4
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr4_-_63154130 | 0.91 |
ENSMUST00000030041.4
|
Ambp
|
alpha 1 microglobulin/bikunin |
| chr2_-_156887172 | 0.89 |
ENSMUST00000109561.3
|
Sla2
|
Src-like-adaptor 2 |
| chr8_-_129065488 | 0.87 |
ENSMUST00000125112.1
ENSMUST00000108747.2 ENSMUST00000095158.4 |
Ccdc7
|
coiled-coil domain containing 7 |
| chr5_-_24423516 | 0.85 |
ENSMUST00000030814.6
|
Cdk5
|
cyclin-dependent kinase 5 |
| chr5_-_138155694 | 0.84 |
ENSMUST00000132318.1
ENSMUST00000049393.8 |
Zfp113
|
zinc finger protein 113 |
| chr11_+_87663087 | 0.84 |
ENSMUST00000165679.1
|
Rnf43
|
ring finger protein 43 |
| chr14_-_49525840 | 0.83 |
ENSMUST00000138884.1
ENSMUST00000074368.4 ENSMUST00000123534.1 |
Slc35f4
|
solute carrier family 35, member F4 |
| chr11_+_80428598 | 0.82 |
ENSMUST00000173938.1
ENSMUST00000017572.7 |
Psmd11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr19_+_4855129 | 0.82 |
ENSMUST00000119694.1
|
Ctsf
|
cathepsin F |
| chr18_-_35655185 | 0.81 |
ENSMUST00000097619.1
|
Prob1
|
proline rich basic protein 1 |
| chr13_+_42709482 | 0.81 |
ENSMUST00000066928.5
ENSMUST00000148891.1 |
Phactr1
|
phosphatase and actin regulator 1 |
| chr5_+_31116722 | 0.80 |
ENSMUST00000114637.1
|
Trim54
|
tripartite motif-containing 54 |
| chr16_-_62847008 | 0.79 |
ENSMUST00000089289.5
|
Arl13b
|
ADP-ribosylation factor-like 13B |
| chr3_+_95282897 | 0.79 |
ENSMUST00000039537.7
ENSMUST00000107187.2 |
Fam63a
|
family with sequence similarity 63, member A |
| chr15_-_41869703 | 0.79 |
ENSMUST00000054742.5
|
Abra
|
actin-binding Rho activating protein |
| chr2_-_76982455 | 0.78 |
ENSMUST00000011934.5
ENSMUST00000099981.2 ENSMUST00000099980.3 ENSMUST00000111882.2 ENSMUST00000140091.1 |
Ttn
|
titin |
| chr12_-_87919857 | 0.77 |
ENSMUST00000180053.1
|
Gm2035
|
predicted pseudogene 2035 |
| chrX_+_150594420 | 0.77 |
ENSMUST00000112713.2
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr2_-_38926217 | 0.77 |
ENSMUST00000076275.4
ENSMUST00000142130.1 |
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr19_+_42255704 | 0.76 |
ENSMUST00000087123.5
|
Golga7b
|
golgi autoantigen, golgin subfamily a, 7B |
| chr13_+_84222286 | 0.75 |
ENSMUST00000057495.8
|
Tmem161b
|
transmembrane protein 161B |
| chr3_-_84270782 | 0.75 |
ENSMUST00000054990.4
|
Trim2
|
tripartite motif-containing 2 |
| chr10_-_24109582 | 0.74 |
ENSMUST00000041180.5
|
Taar9
|
trace amine-associated receptor 9 |
| chr13_-_54611332 | 0.74 |
ENSMUST00000091609.4
|
Cltb
|
clathrin, light polypeptide (Lcb) |
| chr15_-_76232554 | 0.74 |
ENSMUST00000166428.1
|
Plec
|
plectin |
| chr9_-_114844090 | 0.73 |
ENSMUST00000047013.3
|
Cmtm8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr12_-_85824506 | 0.73 |
ENSMUST00000021676.5
ENSMUST00000142331.1 |
0610007P14Rik
|
RIKEN cDNA 0610007P14 gene |
| chr3_-_146770603 | 0.73 |
ENSMUST00000106138.1
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr13_-_31559333 | 0.72 |
ENSMUST00000170573.1
|
A530084C06Rik
|
RIKEN cDNA A530084C06 gene |
| chr4_+_138454305 | 0.71 |
ENSMUST00000050918.3
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr11_+_76202007 | 0.71 |
ENSMUST00000094014.3
|
Fam57a
|
family with sequence similarity 57, member A |
| chr15_-_100687908 | 0.71 |
ENSMUST00000023775.7
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr9_+_103112072 | 0.71 |
ENSMUST00000035155.6
|
Rab6b
|
RAB6B, member RAS oncogene family |
| chr16_+_17331371 | 0.70 |
ENSMUST00000023450.6
ENSMUST00000161034.1 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr18_+_5593566 | 0.70 |
ENSMUST00000160910.1
|
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr9_-_63399216 | 0.70 |
ENSMUST00000168665.1
|
2300009A05Rik
|
RIKEN cDNA 2300009A05 gene |
| chr14_+_120478443 | 0.68 |
ENSMUST00000062117.6
|
Rap2a
|
RAS related protein 2a |
| chr2_+_126034647 | 0.67 |
ENSMUST00000064794.7
|
Fgf7
|
fibroblast growth factor 7 |
| chr2_-_26092149 | 0.67 |
ENSMUST00000114159.2
|
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr12_+_80644212 | 0.67 |
ENSMUST00000085245.5
|
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr14_-_26534870 | 0.67 |
ENSMUST00000139075.1
ENSMUST00000102956.1 |
Slmap
|
sarcolemma associated protein |
| chr19_+_39060998 | 0.66 |
ENSMUST00000087236.4
|
Cyp2c65
|
cytochrome P450, family 2, subfamily c, polypeptide 65 |
| chr12_+_76081645 | 0.66 |
ENSMUST00000154509.1
|
Syne2
|
spectrin repeat containing, nuclear envelope 2 |
| chr10_+_86705811 | 0.66 |
ENSMUST00000061458.7
ENSMUST00000075632.6 |
BC030307
|
cDNA sequence BC030307 |
| chr11_+_76202084 | 0.66 |
ENSMUST00000169560.1
|
Fam57a
|
family with sequence similarity 57, member A |
| chr3_-_79145875 | 0.65 |
ENSMUST00000118340.1
|
Rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr2_-_130397525 | 0.65 |
ENSMUST00000028897.7
|
Cpxm1
|
carboxypeptidase X 1 (M14 family) |
| chr4_-_118690463 | 0.65 |
ENSMUST00000060562.3
|
Olfr1342
|
olfactory receptor 1342 |
| chr1_-_74749221 | 0.65 |
ENSMUST00000081636.6
|
Prkag3
|
protein kinase, AMP-activated, gamma 3 non-catatlytic subunit |
| chr16_+_20672716 | 0.64 |
ENSMUST00000044783.7
ENSMUST00000115463.1 ENSMUST00000142344.1 ENSMUST00000073840.5 ENSMUST00000140576.1 ENSMUST00000115457.1 |
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr11_-_120630516 | 0.64 |
ENSMUST00000106181.1
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chrX_-_18461371 | 0.63 |
ENSMUST00000044188.4
|
4930578C19Rik
|
RIKEN cDNA 4930578C19 gene |
| chr17_-_35172608 | 0.63 |
ENSMUST00000173106.1
|
Aif1
|
allograft inflammatory factor 1 |
| chr9_+_104002546 | 0.62 |
ENSMUST00000035167.8
ENSMUST00000117054.1 |
Nphp3
|
nephronophthisis 3 (adolescent) |
| chr10_+_24076500 | 0.62 |
ENSMUST00000051133.5
|
Taar8a
|
trace amine-associated receptor 8A |
| chr5_-_30196418 | 0.62 |
ENSMUST00000125367.1
|
Gpr113
|
G protein-coupled receptor 113 |
| chr16_-_88563166 | 0.62 |
ENSMUST00000049697.4
|
Cldn8
|
claudin 8 |
| chr6_+_110645572 | 0.61 |
ENSMUST00000071076.6
ENSMUST00000172951.1 |
Grm7
|
glutamate receptor, metabotropic 7 |
| chr16_+_20548577 | 0.61 |
ENSMUST00000003319.5
|
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr16_-_36990449 | 0.60 |
ENSMUST00000075869.6
|
Fbxo40
|
F-box protein 40 |
| chr10_-_14718191 | 0.60 |
ENSMUST00000020016.4
|
Gje1
|
gap junction protein, epsilon 1 |
| chr2_-_146511899 | 0.59 |
ENSMUST00000131824.1
|
Ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr12_+_30584429 | 0.58 |
ENSMUST00000057151.8
|
Tmem18
|
transmembrane protein 18 |
| chr15_+_25622525 | 0.58 |
ENSMUST00000110457.1
ENSMUST00000137601.1 |
Myo10
|
myosin X |
| chr8_-_11478618 | 0.58 |
ENSMUST00000033900.5
|
Rab20
|
RAB20, member RAS oncogene family |
| chr6_+_21985903 | 0.58 |
ENSMUST00000137437.1
ENSMUST00000115383.2 |
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr5_-_125294107 | 0.58 |
ENSMUST00000127148.1
|
Scarb1
|
scavenger receptor class B, member 1 |
| chr1_+_127306706 | 0.58 |
ENSMUST00000171405.1
|
Mgat5
|
mannoside acetylglucosaminyltransferase 5 |
| chr1_+_87124946 | 0.57 |
ENSMUST00000044878.3
|
Akp3
|
alkaline phosphatase 3, intestine, not Mn requiring |
| chr9_+_45055211 | 0.57 |
ENSMUST00000114663.2
|
Mpzl3
|
myelin protein zero-like 3 |
| chr14_-_54253907 | 0.56 |
ENSMUST00000128231.1
|
Dad1
|
defender against cell death 1 |
| chr9_+_45055166 | 0.56 |
ENSMUST00000114664.1
ENSMUST00000093856.3 |
Mpzl3
|
myelin protein zero-like 3 |
| chrX_+_93183227 | 0.55 |
ENSMUST00000088133.3
|
BC061195
|
cDNA sequence BC061195 |
| chr12_+_35047180 | 0.55 |
ENSMUST00000048519.9
ENSMUST00000163677.1 |
Snx13
|
sorting nexin 13 |
| chr11_-_97115327 | 0.55 |
ENSMUST00000001484.2
|
Tbx21
|
T-box 21 |
| chr15_-_85503227 | 0.55 |
ENSMUST00000178942.1
|
7530416G11Rik
|
RIKEN cDNA 7530416G11 gene |
| chrX_+_7638674 | 0.54 |
ENSMUST00000128890.1
|
Syp
|
synaptophysin |
| chr6_+_41538218 | 0.54 |
ENSMUST00000103291.1
|
Trbc1
|
T cell receptor beta, constant region 1 |
| chr2_-_30093642 | 0.54 |
ENSMUST00000102865.4
|
Zdhhc12
|
zinc finger, DHHC domain containing 12 |
| chr15_-_67113909 | 0.54 |
ENSMUST00000092640.5
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr4_+_155839724 | 0.53 |
ENSMUST00000030947.3
|
Mxra8
|
matrix-remodelling associated 8 |
| chr12_-_113361232 | 0.53 |
ENSMUST00000103423.1
|
Ighg3
|
Immunoglobulin heavy constant gamma 3 |
| chr10_-_42583628 | 0.53 |
ENSMUST00000019938.4
|
Nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr6_+_21986438 | 0.52 |
ENSMUST00000115382.1
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr13_-_67332525 | 0.52 |
ENSMUST00000168892.1
ENSMUST00000109735.2 |
Zfp595
|
zinc finger protein 595 |
| chr2_-_167188787 | 0.52 |
ENSMUST00000059826.8
|
Kcnb1
|
potassium voltage gated channel, Shab-related subfamily, member 1 |
| chr4_+_59003121 | 0.52 |
ENSMUST00000095070.3
ENSMUST00000174664.1 |
Dnajc25
Gm20503
|
DnaJ (Hsp40) homolog, subfamily C, member 25 predicted gene 20503 |
| chr9_-_57606234 | 0.52 |
ENSMUST00000045068.8
|
Cplx3
|
complexin 3 |
| chr11_+_82035569 | 0.52 |
ENSMUST00000000193.5
|
Ccl2
|
chemokine (C-C motif) ligand 2 |
| chr10_+_69213084 | 0.51 |
ENSMUST00000163497.1
ENSMUST00000164212.1 ENSMUST00000067908.7 |
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr1_-_163725123 | 0.50 |
ENSMUST00000159679.1
|
Mettl11b
|
methyltransferase like 11B |
| chr9_-_44965519 | 0.50 |
ENSMUST00000125642.1
ENSMUST00000117506.1 ENSMUST00000117549.1 |
Ube4a
|
ubiquitination factor E4A, UFD2 homolog (S. cerevisiae) |
| chr14_-_34588607 | 0.50 |
ENSMUST00000090040.4
|
Ldb3
|
LIM domain binding 3 |
| chr4_-_57300751 | 0.50 |
ENSMUST00000151964.1
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr6_-_39118211 | 0.50 |
ENSMUST00000038398.6
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr15_+_83779999 | 0.49 |
ENSMUST00000046168.5
|
Mpped1
|
metallophosphoesterase domain containing 1 |
| chr7_-_45136102 | 0.49 |
ENSMUST00000125500.1
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr8_+_46739745 | 0.49 |
ENSMUST00000034041.7
|
Irf2
|
interferon regulatory factor 2 |
| chr8_+_44950208 | 0.48 |
ENSMUST00000098796.3
|
Fat1
|
FAT tumor suppressor homolog 1 (Drosophila) |
| chr9_-_87731248 | 0.48 |
ENSMUST00000034991.7
|
Tbx18
|
T-box18 |
| chr16_+_13358375 | 0.48 |
ENSMUST00000149359.1
|
Mkl2
|
MKL/myocardin-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Maf_Nrl
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 46.7 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 2.4 | 9.4 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 1.2 | 13.6 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.8 | 5.6 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.8 | 1.6 | GO:0046271 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) phenylpropanoid catabolic process(GO:0046271) |
| 0.7 | 1.4 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.7 | 2.6 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.6 | 1.7 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.6 | 9.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.5 | 6.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.5 | 0.5 | GO:0002436 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.5 | 2.4 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.5 | 1.9 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.5 | 6.5 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.4 | 2.5 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.4 | 1.2 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.3 | 1.3 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.3 | 1.8 | GO:0060523 | Sertoli cell proliferation(GO:0060011) prostate epithelial cord elongation(GO:0060523) |
| 0.3 | 5.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.3 | 0.9 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.3 | 1.2 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.3 | 2.0 | GO:0001878 | response to yeast(GO:0001878) |
| 0.3 | 0.9 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.3 | 1.7 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.3 | 1.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.3 | 1.4 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) |
| 0.3 | 1.9 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.3 | 1.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.2 | 1.2 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.2 | 0.5 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.2 | 0.7 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.2 | 2.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 0.7 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.2 | 1.7 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.2 | 1.5 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.2 | 4.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.2 | 1.2 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.2 | 2.0 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 0.6 | GO:0015920 | regulation of phosphatidylcholine catabolic process(GO:0010899) lipopolysaccharide transport(GO:0015920) |
| 0.2 | 0.9 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 0.5 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.2 | 1.6 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 1.1 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.2 | 0.6 | GO:0014900 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.1 | 0.3 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 | 0.4 | GO:0060364 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 0.1 | 0.4 | GO:0051659 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.8 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.5 | GO:0060857 | establishment of glial blood-brain barrier(GO:0060857) |
| 0.1 | 0.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.7 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 2.7 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 0.6 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 1.4 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 2.0 | GO:0051608 | histamine transport(GO:0051608) |
| 0.1 | 0.6 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 1.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.6 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.4 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 | 1.9 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.1 | 0.2 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.1 | 0.3 | GO:2000978 | trochlear nerve development(GO:0021558) auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.1 | 0.7 | GO:0051541 | elastin metabolic process(GO:0051541) pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.8 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.1 | 0.6 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 0.6 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 0.3 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 0.1 | 0.7 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.4 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.5 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.4 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.1 | 1.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.9 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.4 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 1.7 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.7 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.3 | GO:0070537 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.7 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 0.8 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 1.1 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.7 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.5 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.1 | 1.1 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 0.2 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.1 | 0.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.6 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.5 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.6 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.2 | GO:0060128 | otic vesicle formation(GO:0030916) corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.5 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.2 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 1.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 1.7 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 6.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.3 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.2 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.4 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 1.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 1.0 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.5 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.3 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.0 | 0.4 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.1 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0021995 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 0.2 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.2 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 4.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.6 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.4 | GO:0051043 | regulation of membrane protein ectodomain proteolysis(GO:0051043) |
| 0.0 | 0.7 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.6 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.0 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.5 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.4 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 1.7 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.2 | GO:1990000 | amyloid fibril formation(GO:1990000) |
| 0.0 | 0.1 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 | 0.3 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 1.3 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.8 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.6 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 46.7 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) insulin-like growth factor ternary complex(GO:0042567) |
| 1.0 | 10.7 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.8 | 9.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 1.2 | GO:0044753 | amphisome(GO:0044753) |
| 0.3 | 0.9 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.3 | 1.8 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.3 | 0.8 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 1.2 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.2 | 1.7 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 0.6 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.2 | 1.0 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 1.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.5 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 2.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 1.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.4 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 2.0 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.7 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 1.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.9 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.7 | GO:0034992 | lamellipodium membrane(GO:0031258) microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.8 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 1.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.5 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 0.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 0.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.8 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 2.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.6 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.5 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 2.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.8 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 3.4 | 13.6 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 2.7 | 10.7 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.2 | 46.7 | GO:0043539 | insulin-like growth factor receptor binding(GO:0005159) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.7 | 2.0 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.6 | 5.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.6 | 1.8 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.5 | 2.5 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.4 | 1.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.4 | 1.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.4 | 1.5 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.3 | 2.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 1.7 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.3 | 0.9 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.3 | 1.9 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 2.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 5.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 2.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.2 | 0.6 | GO:0070905 | serine binding(GO:0070905) |
| 0.2 | 1.6 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.2 | 1.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 1.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 1.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 1.1 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.2 | 7.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.6 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.6 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.4 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 0.7 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.7 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.1 | 4.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.5 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 1.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.3 | GO:0071820 | N-box binding(GO:0071820) |
| 0.1 | 0.4 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.4 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 0.8 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.0 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 1.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.9 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 3.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.4 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 0.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.9 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.2 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.1 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 5.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.6 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 1.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.2 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 1.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 2.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 6.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 2.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.7 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 1.4 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 1.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0016918 | retinal binding(GO:0016918) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 46.9 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 4.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 3.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 2.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 5.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 3.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.6 | 5.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.5 | 11.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 9.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 4.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 2.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 4.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 2.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 3.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 1.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.9 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 1.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 2.1 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 1.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.6 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |