Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Klf15
Z-value: 1.30
Transcription factors associated with Klf15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf15
|
ENSMUSG00000030087.5 | Kruppel-like factor 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf15 | mm10_v2_chr6_+_90462562_90462587 | 0.65 | 1.9e-05 | Click! |
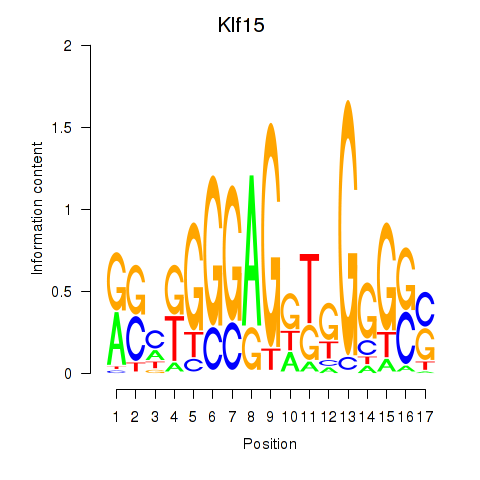
Activity profile of Klf15 motif
Sorted Z-values of Klf15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_148045891 | 5.11 |
ENSMUST00000109964.1
|
Foxa2
|
forkhead box A2 |
| chr5_+_130448801 | 3.91 |
ENSMUST00000111288.2
|
Caln1
|
calneuron 1 |
| chr18_-_38211957 | 3.45 |
ENSMUST00000159405.1
ENSMUST00000160721.1 |
Pcdh1
|
protocadherin 1 |
| chr11_+_98348404 | 3.23 |
ENSMUST00000078694.6
|
Ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr7_-_99626936 | 3.08 |
ENSMUST00000178124.1
|
Gm4980
|
predicted gene 4980 |
| chr4_+_97777780 | 2.98 |
ENSMUST00000107062.2
ENSMUST00000052018.5 ENSMUST00000107057.1 |
Nfia
|
nuclear factor I/A |
| chr2_-_25501717 | 2.91 |
ENSMUST00000015227.3
|
C8g
|
complement component 8, gamma polypeptide |
| chr8_-_41133697 | 2.89 |
ENSMUST00000155055.1
ENSMUST00000059115.6 ENSMUST00000145860.1 |
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr2_-_5714490 | 2.73 |
ENSMUST00000044009.7
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr10_-_81430966 | 2.58 |
ENSMUST00000117966.1
|
Nfic
|
nuclear factor I/C |
| chr4_+_139622842 | 2.55 |
ENSMUST00000039818.9
|
Aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr2_-_130642770 | 2.53 |
ENSMUST00000045761.6
|
Lzts3
|
leucine zipper, putative tumor suppressor family member 3 |
| chrX_-_136868537 | 2.40 |
ENSMUST00000058814.6
|
Rab9b
|
RAB9B, member RAS oncogene family |
| chr9_-_97018823 | 2.31 |
ENSMUST00000055433.4
|
Spsb4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr16_+_44173239 | 2.26 |
ENSMUST00000119746.1
|
Gm608
|
predicted gene 608 |
| chr10_-_127666598 | 2.25 |
ENSMUST00000099157.3
|
Nab2
|
Ngfi-A binding protein 2 |
| chr12_+_8771405 | 2.24 |
ENSMUST00000171158.1
|
Sdc1
|
syndecan 1 |
| chr4_+_83525540 | 2.24 |
ENSMUST00000053414.6
ENSMUST00000126429.1 |
Ccdc171
|
coiled-coil domain containing 171 |
| chr9_+_46012810 | 2.23 |
ENSMUST00000126865.1
|
Sik3
|
SIK family kinase 3 |
| chr7_-_140102367 | 2.21 |
ENSMUST00000142105.1
|
Fuom
|
fucose mutarotase |
| chr19_+_6497772 | 2.17 |
ENSMUST00000113458.1
ENSMUST00000113459.1 |
Nrxn2
|
neurexin II |
| chrX_+_161717498 | 2.13 |
ENSMUST00000061514.7
|
Rai2
|
retinoic acid induced 2 |
| chr5_+_30588078 | 2.12 |
ENSMUST00000066295.2
|
Kcnk3
|
potassium channel, subfamily K, member 3 |
| chr9_-_21927515 | 2.11 |
ENSMUST00000178988.1
ENSMUST00000046831.9 |
Tmem205
|
transmembrane protein 205 |
| chr19_+_53903351 | 2.11 |
ENSMUST00000025931.6
ENSMUST00000165617.1 |
Pdcd4
|
programmed cell death 4 |
| chr7_+_26307190 | 2.09 |
ENSMUST00000098657.3
|
Cyp2a4
|
cytochrome P450, family 2, subfamily a, polypeptide 4 |
| chr12_+_8771317 | 2.07 |
ENSMUST00000020911.7
|
Sdc1
|
syndecan 1 |
| chr11_-_100770926 | 1.85 |
ENSMUST00000139341.1
ENSMUST00000017891.7 |
Ghdc
|
GH3 domain containing |
| chr7_-_80403315 | 1.84 |
ENSMUST00000147150.1
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr10_-_127666673 | 1.80 |
ENSMUST00000026469.2
|
Nab2
|
Ngfi-A binding protein 2 |
| chr9_+_21927471 | 1.78 |
ENSMUST00000170304.1
ENSMUST00000006403.6 |
Ccdc159
|
coiled-coil domain containing 159 |
| chr7_-_140102326 | 1.74 |
ENSMUST00000128527.1
|
Fuom
|
fucose mutarotase |
| chr2_+_140395446 | 1.62 |
ENSMUST00000110061.1
|
Macrod2
|
MACRO domain containing 2 |
| chr4_-_129239165 | 1.55 |
ENSMUST00000097873.3
|
C77080
|
expressed sequence C77080 |
| chr5_+_148959263 | 1.54 |
ENSMUST00000135240.1
|
Gm15409
|
predicted gene 15409 |
| chr15_+_57694651 | 1.54 |
ENSMUST00000096430.4
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr2_-_90580578 | 1.46 |
ENSMUST00000168621.2
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr15_+_87625214 | 1.44 |
ENSMUST00000068088.6
|
Fam19a5
|
family with sequence similarity 19, member A5 |
| chr7_-_80402743 | 1.44 |
ENSMUST00000122232.1
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr8_+_111536492 | 1.43 |
ENSMUST00000168428.1
ENSMUST00000171182.1 |
Znrf1
|
zinc and ring finger 1 |
| chr9_+_46012822 | 1.42 |
ENSMUST00000120463.2
ENSMUST00000120247.1 |
Sik3
|
SIK family kinase 3 |
| chr4_-_148159571 | 1.40 |
ENSMUST00000167160.1
ENSMUST00000151246.1 |
Fbxo44
|
F-box protein 44 |
| chr10_-_54075702 | 1.39 |
ENSMUST00000105470.1
|
Man1a
|
mannosidase 1, alpha |
| chr10_-_59616667 | 1.39 |
ENSMUST00000020312.6
|
Mcu
|
mitochondrial calcium uniporter |
| chr5_+_127632238 | 1.38 |
ENSMUST00000118139.1
|
Glt1d1
|
glycosyltransferase 1 domain containing 1 |
| chr10_-_54075730 | 1.37 |
ENSMUST00000105469.1
ENSMUST00000003843.8 |
Man1a
|
mannosidase 1, alpha |
| chr1_+_59482133 | 1.36 |
ENSMUST00000114246.2
ENSMUST00000037105.6 |
Fzd7
|
frizzled homolog 7 (Drosophila) |
| chr11_-_97574040 | 1.35 |
ENSMUST00000107593.1
|
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr16_-_46496772 | 1.35 |
ENSMUST00000149901.1
ENSMUST00000096052.2 |
Pvrl3
|
poliovirus receptor-related 3 |
| chr2_+_140395309 | 1.35 |
ENSMUST00000110067.1
ENSMUST00000110064.1 ENSMUST00000110063.1 ENSMUST00000110062.1 ENSMUST00000078027.5 ENSMUST00000043836.7 |
Macrod2
|
MACRO domain containing 2 |
| chr2_+_32606979 | 1.33 |
ENSMUST00000113289.1
ENSMUST00000095044.3 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr13_+_58402546 | 1.31 |
ENSMUST00000042450.8
|
Rmi1
|
RMI1, RecQ mediated genome instability 1, homolog (S. cerevisiae) |
| chr4_-_57300362 | 1.30 |
ENSMUST00000153926.1
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr16_-_96082389 | 1.28 |
ENSMUST00000099502.2
ENSMUST00000023631.8 ENSMUST00000113829.1 ENSMUST00000153398.1 |
Brwd1
|
bromodomain and WD repeat domain containing 1 |
| chr12_+_41024090 | 1.28 |
ENSMUST00000132121.1
|
Immp2l
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr11_-_3863895 | 1.28 |
ENSMUST00000070552.7
|
Osbp2
|
oxysterol binding protein 2 |
| chrX_+_141475385 | 1.27 |
ENSMUST00000112931.1
ENSMUST00000112930.1 |
Col4a5
|
collagen, type IV, alpha 5 |
| chr17_+_43801823 | 1.26 |
ENSMUST00000044895.5
|
Rcan2
|
regulator of calcineurin 2 |
| chr2_-_120314141 | 1.26 |
ENSMUST00000054651.7
|
Pla2g4f
|
phospholipase A2, group IVF |
| chr7_+_28180226 | 1.25 |
ENSMUST00000172467.1
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr17_+_80944611 | 1.24 |
ENSMUST00000025092.4
|
Tmem178
|
transmembrane protein 178 |
| chr16_+_20733104 | 1.18 |
ENSMUST00000115423.1
ENSMUST00000007171.6 |
Chrd
|
chordin |
| chr17_-_32947372 | 1.17 |
ENSMUST00000139353.1
|
Cyp4f13
|
cytochrome P450, family 4, subfamily f, polypeptide 13 |
| chr17_-_15375969 | 1.16 |
ENSMUST00000014917.7
|
Dll1
|
delta-like 1 (Drosophila) |
| chr1_-_161034794 | 1.16 |
ENSMUST00000177003.1
ENSMUST00000162226.2 ENSMUST00000159250.2 |
Zbtb37
|
zinc finger and BTB domain containing 37 |
| chr11_-_97573929 | 1.15 |
ENSMUST00000126287.1
ENSMUST00000107590.1 |
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr11_-_72796028 | 1.15 |
ENSMUST00000156294.1
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr3_+_117575268 | 1.14 |
ENSMUST00000039564.6
|
4833424O15Rik
|
RIKEN cDNA 4833424O15 gene |
| chr17_-_32947389 | 1.14 |
ENSMUST00000075253.6
|
Cyp4f13
|
cytochrome P450, family 4, subfamily f, polypeptide 13 |
| chr4_+_97777606 | 1.14 |
ENSMUST00000075448.6
ENSMUST00000092532.6 |
Nfia
|
nuclear factor I/A |
| chr2_-_173276144 | 1.13 |
ENSMUST00000139306.1
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr7_-_140102384 | 1.12 |
ENSMUST00000120034.1
ENSMUST00000121115.1 ENSMUST00000026539.7 |
Fuom
|
fucose mutarotase |
| chr1_+_36511867 | 1.11 |
ENSMUST00000001166.7
ENSMUST00000097776.3 |
Cnnm3
|
cyclin M3 |
| chr19_-_43674844 | 1.11 |
ENSMUST00000046038.7
|
Slc25a28
|
solute carrier family 25, member 28 |
| chr1_-_93478785 | 1.09 |
ENSMUST00000170883.1
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr8_+_12385769 | 1.09 |
ENSMUST00000080795.8
|
Gm5607
|
predicted gene 5607 |
| chr2_+_18677002 | 1.09 |
ENSMUST00000028071.6
|
Bmi1
|
Bmi1 polycomb ring finger oncogene |
| chr13_-_29984219 | 1.08 |
ENSMUST00000146092.1
|
E2f3
|
E2F transcription factor 3 |
| chr19_-_44029201 | 1.08 |
ENSMUST00000026211.8
|
Cyp2c44
|
cytochrome P450, family 2, subfamily c, polypeptide 44 |
| chr2_+_69897220 | 1.08 |
ENSMUST00000055758.9
ENSMUST00000112251.2 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr14_-_70520254 | 1.07 |
ENSMUST00000022693.7
|
Bmp1
|
bone morphogenetic protein 1 |
| chr16_-_46496955 | 1.02 |
ENSMUST00000023335.6
ENSMUST00000023334.8 |
Pvrl3
|
poliovirus receptor-related 3 |
| chr15_+_25414175 | 1.01 |
ENSMUST00000069992.5
|
Gm5468
|
predicted gene 5468 |
| chr19_-_29812952 | 1.00 |
ENSMUST00000099525.3
|
Ranbp6
|
RAN binding protein 6 |
| chr14_-_39472825 | 0.99 |
ENSMUST00000168810.2
ENSMUST00000173780.1 ENSMUST00000166968.2 |
Nrg3
|
neuregulin 3 |
| chr17_-_70851189 | 0.99 |
ENSMUST00000059775.8
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr2_+_31314303 | 0.98 |
ENSMUST00000113532.2
|
Hmcn2
|
hemicentin 2 |
| chr9_+_60794468 | 0.97 |
ENSMUST00000050183.6
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr11_-_100759740 | 0.97 |
ENSMUST00000107361.2
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr9_+_108080436 | 0.96 |
ENSMUST00000035211.7
ENSMUST00000162886.1 |
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr7_+_28180272 | 0.94 |
ENSMUST00000173223.1
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr4_+_107178399 | 0.93 |
ENSMUST00000030361.4
ENSMUST00000128123.1 ENSMUST00000106753.1 |
Tmem59
|
transmembrane protein 59 |
| chr2_+_91257323 | 0.93 |
ENSMUST00000111349.2
ENSMUST00000131711.1 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr7_+_100706623 | 0.93 |
ENSMUST00000107042.1
|
Fam168a
|
family with sequence similarity 168, member A |
| chr11_+_69765970 | 0.89 |
ENSMUST00000108642.1
ENSMUST00000156932.1 |
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr2_+_18064564 | 0.89 |
ENSMUST00000114671.1
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr10_-_7956223 | 0.87 |
ENSMUST00000146444.1
|
Tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr8_-_91134027 | 0.86 |
ENSMUST00000125257.1
|
Aktip
|
thymoma viral proto-oncogene 1 interacting protein |
| chr4_-_124850670 | 0.86 |
ENSMUST00000163946.1
ENSMUST00000106190.3 |
1110065P20Rik
|
RIKEN cDNA 1110065P20 gene |
| chr12_-_98737405 | 0.85 |
ENSMUST00000170188.1
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr13_-_103920508 | 0.85 |
ENSMUST00000053927.5
ENSMUST00000091269.4 ENSMUST00000022222.5 |
Erbb2ip
|
Erbb2 interacting protein |
| chr7_+_100706702 | 0.84 |
ENSMUST00000049053.7
|
Fam168a
|
family with sequence similarity 168, member A |
| chr12_+_108792946 | 0.84 |
ENSMUST00000021692.7
|
Yy1
|
YY1 transcription factor |
| chr15_+_102921103 | 0.84 |
ENSMUST00000001700.6
|
Hoxc13
|
homeobox C13 |
| chr17_+_46383725 | 0.83 |
ENSMUST00000113481.1
ENSMUST00000138127.1 |
Zfp318
|
zinc finger protein 318 |
| chr8_-_93810225 | 0.83 |
ENSMUST00000181864.1
|
Gm26843
|
predicted gene, 26843 |
| chr13_-_103920295 | 0.83 |
ENSMUST00000169083.1
|
Erbb2ip
|
Erbb2 interacting protein |
| chr16_-_96082513 | 0.83 |
ENSMUST00000113827.1
|
Brwd1
|
bromodomain and WD repeat domain containing 1 |
| chr13_+_51100810 | 0.81 |
ENSMUST00000095797.5
|
Spin1
|
spindlin 1 |
| chr8_-_91133942 | 0.81 |
ENSMUST00000120213.1
ENSMUST00000109609.2 |
Aktip
|
thymoma viral proto-oncogene 1 interacting protein |
| chr18_+_36281069 | 0.80 |
ENSMUST00000051301.3
|
Pura
|
purine rich element binding protein A |
| chr4_+_122995944 | 0.79 |
ENSMUST00000106252.2
|
Mycl
|
v-myc myelocytomatosis viral oncogene homolog, lung carcinoma derived (avian) |
| chr4_+_122996035 | 0.79 |
ENSMUST00000030407.7
|
Mycl
|
v-myc myelocytomatosis viral oncogene homolog, lung carcinoma derived (avian) |
| chr11_+_50602072 | 0.78 |
ENSMUST00000040523.8
|
Adamts2
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 2 |
| chr5_+_135064206 | 0.77 |
ENSMUST00000071263.5
|
Dnajc30
|
DnaJ (Hsp40) homolog, subfamily C, member 30 |
| chr3_-_139075178 | 0.76 |
ENSMUST00000098574.2
ENSMUST00000029796.6 |
Rap1gds1
|
RAP1, GTP-GDP dissociation stimulator 1 |
| chr4_-_124850473 | 0.76 |
ENSMUST00000137769.2
|
1110065P20Rik
|
RIKEN cDNA 1110065P20 gene |
| chr16_-_84835484 | 0.75 |
ENSMUST00000114191.1
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr16_+_20673264 | 0.75 |
ENSMUST00000154950.1
ENSMUST00000115461.1 |
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr3_+_121426495 | 0.74 |
ENSMUST00000029773.8
|
Cnn3
|
calponin 3, acidic |
| chr1_-_168432270 | 0.74 |
ENSMUST00000072863.4
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr9_+_119402444 | 0.74 |
ENSMUST00000035093.8
ENSMUST00000165044.1 |
Acvr2b
|
activin receptor IIB |
| chr2_+_18064645 | 0.73 |
ENSMUST00000114680.2
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr1_-_168431695 | 0.72 |
ENSMUST00000176790.1
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr1_-_56971762 | 0.71 |
ENSMUST00000114415.3
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr9_+_65630552 | 0.70 |
ENSMUST00000055844.8
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr11_-_116307168 | 0.70 |
ENSMUST00000124281.1
|
Exoc7
|
exocyst complex component 7 |
| chr10_+_39732099 | 0.70 |
ENSMUST00000019986.6
|
Rev3l
|
REV3-like, catalytic subunit of DNA polymerase zeta RAD54 like (S. cerevisiae) |
| chr4_-_126533472 | 0.69 |
ENSMUST00000084289.4
|
Ago4
|
argonaute RISC catalytic subunit 4 |
| chr2_-_65238573 | 0.68 |
ENSMUST00000090896.3
ENSMUST00000155082.1 |
Cobll1
|
Cobl-like 1 |
| chr9_+_25481547 | 0.68 |
ENSMUST00000040677.5
|
Eepd1
|
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chr4_+_131921771 | 0.67 |
ENSMUST00000094666.3
|
Tmem200b
|
transmembrane protein 200B |
| chr2_-_20968881 | 0.66 |
ENSMUST00000114594.1
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr10_-_84440591 | 0.66 |
ENSMUST00000020220.8
|
Nuak1
|
NUAK family, SNF1-like kinase, 1 |
| chr18_-_31949571 | 0.66 |
ENSMUST00000064016.5
|
Gpr17
|
G protein-coupled receptor 17 |
| chr11_-_102296618 | 0.66 |
ENSMUST00000107132.2
ENSMUST00000073234.2 |
Atxn7l3
|
ataxin 7-like 3 |
| chr1_-_16519284 | 0.65 |
ENSMUST00000162751.1
ENSMUST00000027052.6 ENSMUST00000149320.2 |
Stau2
|
staufen (RNA binding protein) homolog 2 (Drosophila) |
| chr9_+_118478182 | 0.64 |
ENSMUST00000111763.1
|
Eomes
|
eomesodermin homolog (Xenopus laevis) |
| chr14_+_70530819 | 0.63 |
ENSMUST00000047331.6
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr16_-_84835557 | 0.63 |
ENSMUST00000138279.1
ENSMUST00000023608.7 |
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr11_+_23306884 | 0.62 |
ENSMUST00000180046.1
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr18_-_39489157 | 0.61 |
ENSMUST00000131885.1
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr11_-_5261558 | 0.60 |
ENSMUST00000020662.8
|
Kremen1
|
kringle containing transmembrane protein 1 |
| chr13_-_9764431 | 0.60 |
ENSMUST00000154994.1
ENSMUST00000146039.1 ENSMUST00000110635.1 ENSMUST00000110638.1 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr9_+_118478344 | 0.60 |
ENSMUST00000035020.8
|
Eomes
|
eomesodermin homolog (Xenopus laevis) |
| chr4_-_124850652 | 0.60 |
ENSMUST00000125776.1
|
1110065P20Rik
|
RIKEN cDNA 1110065P20 gene |
| chrX_-_152368680 | 0.60 |
ENSMUST00000070316.5
|
Gpr173
|
G-protein coupled receptor 173 |
| chr1_-_16519201 | 0.59 |
ENSMUST00000159558.1
ENSMUST00000054668.6 ENSMUST00000162627.1 ENSMUST00000162007.1 ENSMUST00000128957.2 ENSMUST00000115359.3 ENSMUST00000151888.1 |
Stau2
|
staufen (RNA binding protein) homolog 2 (Drosophila) |
| chr19_+_6047081 | 0.59 |
ENSMUST00000025723.8
|
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr5_-_65492984 | 0.57 |
ENSMUST00000139122.1
|
Smim14
|
small integral membrane protein 14 |
| chr12_+_102283116 | 0.57 |
ENSMUST00000101114.4
ENSMUST00000150795.1 |
Rin3
|
Ras and Rab interactor 3 |
| chr7_-_16874845 | 0.57 |
ENSMUST00000181501.1
|
9330104G04Rik
|
RIKEN cDNA 9330104G04 gene |
| chrX_-_20617574 | 0.56 |
ENSMUST00000116621.1
|
Ndufb11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11 |
| chr2_-_28840274 | 0.56 |
ENSMUST00000037117.5
ENSMUST00000171404.1 |
Gtf3c4
|
general transcription factor IIIC, polypeptide 4 |
| chr5_-_106696530 | 0.56 |
ENSMUST00000137285.1
ENSMUST00000124263.1 ENSMUST00000112695.1 ENSMUST00000155495.1 ENSMUST00000135108.1 |
Zfp644
|
zinc finger protein 644 |
| chr7_-_119184374 | 0.56 |
ENSMUST00000084650.4
|
Gpr139
|
G protein-coupled receptor 139 |
| chr10_+_79716588 | 0.56 |
ENSMUST00000099513.1
ENSMUST00000020581.2 |
Hcn2
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 2 |
| chr6_+_8949670 | 0.55 |
ENSMUST00000060369.3
|
Nxph1
|
neurexophilin 1 |
| chr8_-_121652895 | 0.55 |
ENSMUST00000046386.4
|
Zcchc14
|
zinc finger, CCHC domain containing 14 |
| chr19_+_60889749 | 0.54 |
ENSMUST00000003313.8
|
Grk5
|
G protein-coupled receptor kinase 5 |
| chr1_+_105990384 | 0.54 |
ENSMUST00000119166.1
|
Zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr7_+_16310412 | 0.54 |
ENSMUST00000136781.1
|
Bbc3
|
BCL2 binding component 3 |
| chr11_-_116306696 | 0.54 |
ENSMUST00000133468.1
ENSMUST00000106411.3 ENSMUST00000106413.3 ENSMUST00000021147.7 |
Exoc7
|
exocyst complex component 7 |
| chr13_-_71963713 | 0.54 |
ENSMUST00000077337.8
|
Irx1
|
Iroquois related homeobox 1 (Drosophila) |
| chr5_-_135064063 | 0.54 |
ENSMUST00000111205.1
ENSMUST00000141309.1 |
Wbscr22
|
Williams Beuren syndrome chromosome region 22 |
| chr17_+_37050631 | 0.54 |
ENSMUST00000172792.1
ENSMUST00000174347.1 |
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr16_+_35541275 | 0.53 |
ENSMUST00000120756.1
|
Sema5b
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B |
| chr13_-_95223045 | 0.53 |
ENSMUST00000162292.1
|
Pde8b
|
phosphodiesterase 8B |
| chr1_-_87510306 | 0.53 |
ENSMUST00000027477.8
|
Ngef
|
neuronal guanine nucleotide exchange factor |
| chr9_-_59486323 | 0.53 |
ENSMUST00000165322.1
|
Arih1
|
ariadne ubiquitin-conjugating enzyme E2 binding protein homolog 1 (Drosophila) |
| chr17_+_43667389 | 0.52 |
ENSMUST00000170988.1
|
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr6_-_113377712 | 0.52 |
ENSMUST00000113107.1
ENSMUST00000113106.1 |
Tada3
|
transcriptional adaptor 3 |
| chr17_-_22361400 | 0.52 |
ENSMUST00000115535.2
|
Zfp944
|
zinc finger protein 944 |
| chr6_-_113377866 | 0.52 |
ENSMUST00000032410.7
|
Tada3
|
transcriptional adaptor 3 |
| chr2_-_84715160 | 0.52 |
ENSMUST00000035840.5
|
Zdhhc5
|
zinc finger, DHHC domain containing 5 |
| chr10_+_43479140 | 0.51 |
ENSMUST00000167488.1
ENSMUST00000040147.7 |
Bend3
|
BEN domain containing 3 |
| chr13_-_58113592 | 0.51 |
ENSMUST00000160860.1
|
Klhl3
|
kelch-like 3 |
| chr9_-_21592805 | 0.51 |
ENSMUST00000034700.7
ENSMUST00000180365.1 ENSMUST00000078572.7 |
Yipf2
|
Yip1 domain family, member 2 |
| chr11_-_62731865 | 0.51 |
ENSMUST00000150336.1
|
Zfp287
|
zinc finger protein 287 |
| chr18_-_38284391 | 0.50 |
ENSMUST00000025311.5
|
Pcdh12
|
protocadherin 12 |
| chr2_-_153632679 | 0.50 |
ENSMUST00000109782.1
|
Commd7
|
COMM domain containing 7 |
| chr6_-_99666762 | 0.49 |
ENSMUST00000032151.2
|
Eif4e3
|
eukaryotic translation initiation factor 4E member 3 |
| chr17_+_26933070 | 0.49 |
ENSMUST00000073724.5
|
Phf1
|
PHD finger protein 1 |
| chr11_-_116306652 | 0.48 |
ENSMUST00000126731.1
|
Exoc7
|
exocyst complex component 7 |
| chr1_-_124045247 | 0.48 |
ENSMUST00000112603.2
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr7_-_27396542 | 0.48 |
ENSMUST00000108363.1
|
Sptbn4
|
spectrin beta, non-erythrocytic 4 |
| chr17_-_80728026 | 0.46 |
ENSMUST00000112389.2
ENSMUST00000025089.7 |
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr4_+_47353222 | 0.46 |
ENSMUST00000007757.8
|
Tgfbr1
|
transforming growth factor, beta receptor I |
| chr11_+_115420138 | 0.45 |
ENSMUST00000106533.1
ENSMUST00000123345.1 |
Kctd2
|
potassium channel tetramerisation domain containing 2 |
| chr13_-_38151792 | 0.45 |
ENSMUST00000078232.1
|
Gm10129
|
predicted gene 10129 |
| chr17_-_79715034 | 0.44 |
ENSMUST00000024894.1
|
Cyp1b1
|
cytochrome P450, family 1, subfamily b, polypeptide 1 |
| chr18_+_61045139 | 0.44 |
ENSMUST00000025522.4
ENSMUST00000115274.1 |
Pdgfrb
|
platelet derived growth factor receptor, beta polypeptide |
| chr2_+_128818104 | 0.43 |
ENSMUST00000110325.1
|
Tmem87b
|
transmembrane protein 87B |
| chr11_+_115420059 | 0.43 |
ENSMUST00000103035.3
|
Kctd2
|
potassium channel tetramerisation domain containing 2 |
| chr19_+_48206025 | 0.43 |
ENSMUST00000078880.5
|
Sorcs3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr9_+_21592722 | 0.43 |
ENSMUST00000062125.10
|
1810026J23Rik
|
RIKEN cDNA 1810026J23 gene |
| chr3_+_156561950 | 0.42 |
ENSMUST00000041425.5
ENSMUST00000106065.1 |
Negr1
|
neuronal growth regulator 1 |
| chr14_-_55784027 | 0.42 |
ENSMUST00000170223.1
|
Adcy4
|
adenylate cyclase 4 |
| chr1_+_105990652 | 0.42 |
ENSMUST00000118196.1
|
Zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr16_-_34262945 | 0.42 |
ENSMUST00000114953.1
|
Kalrn
|
kalirin, RhoGEF kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Klf15
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 1.4 | 4.3 | GO:0048627 | myoblast development(GO:0048627) |
| 0.8 | 2.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.8 | 3.3 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.8 | 3.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.7 | 2.1 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.6 | 5.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.6 | 1.8 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.6 | 4.0 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.4 | 1.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.4 | 3.0 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.4 | 1.2 | GO:0097101 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel endothelial cell fate specification(GO:0097101) |
| 0.3 | 1.4 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.3 | 1.3 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.3 | 0.9 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.3 | 1.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.3 | 2.7 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.3 | 0.8 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.3 | 1.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 0.2 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.2 | 1.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.2 | 0.7 | GO:1905223 | epicardium morphogenesis(GO:1905223) |
| 0.2 | 1.1 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.2 | 1.2 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.2 | 3.8 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 2.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.5 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.2 | 1.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 1.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.2 | 2.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 1.1 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 1.7 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.2 | 4.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 5.4 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 1.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.4 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 0.4 | GO:0072277 | cell migration involved in vasculogenesis(GO:0035441) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.7 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.1 | 1.6 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 1.3 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.5 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 0.5 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.1 | 0.7 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.5 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 0.4 | GO:0021998 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) regulation of cardiac ventricle development(GO:1904412) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.1 | 1.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.4 | GO:1902868 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 1.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 2.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.7 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 1.0 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.3 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.3 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 | 0.5 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 1.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.6 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 | 0.5 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.4 | GO:0072137 | cell migration involved in endocardial cushion formation(GO:0003273) condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.5 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 3.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.8 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 0.8 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 0.4 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.1 | 1.9 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) |
| 0.1 | 0.4 | GO:0045358 | N-terminal peptidyl-lysine acetylation(GO:0018076) negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.1 | 1.3 | GO:0071236 | cellular response to antibiotic(GO:0071236) |
| 0.1 | 0.4 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 | 2.3 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.1 | 1.8 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.1 | 1.0 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.6 | GO:0071321 | cellular response to cGMP(GO:0071321) potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) regulation of zinc ion transport(GO:0071579) |
| 0.1 | 2.5 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.5 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.1 | 0.7 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.1 | 3.2 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.7 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 1.0 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 1.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.8 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.4 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 0.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.5 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.2 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 0.3 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.1 | 0.7 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 1.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 1.2 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.7 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 1.0 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.2 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 0.6 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.3 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 1.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.9 | GO:0051044 | regulation of membrane protein ectodomain proteolysis(GO:0051043) positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.0 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 2.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.8 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.4 | GO:2000303 | regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 1.7 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 1.1 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 2.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.9 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.3 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.8 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:0032667 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.0 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.6 | GO:1902236 | retrograde protein transport, ER to cytosol(GO:0030970) negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.8 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.5 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.4 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.3 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.8 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.6 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.0 | 0.7 | GO:0017145 | stem cell division(GO:0017145) |
| 0.0 | 0.7 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.3 | 1.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.3 | 2.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 1.0 | GO:0043293 | apoptosome(GO:0043293) |
| 0.2 | 1.7 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 3.7 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.2 | 1.3 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 0.6 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.2 | 1.7 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.6 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 0.9 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.7 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 3.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.5 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.9 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 1.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.3 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.7 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 0.6 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.8 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 0.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 2.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 2.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 2.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 4.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.1 | GO:0042806 | fucose binding(GO:0042806) |
| 1.1 | 3.2 | GO:0031752 | D4 dopamine receptor binding(GO:0031751) D5 dopamine receptor binding(GO:0031752) |
| 0.3 | 2.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 3.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 1.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 2.9 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 0.8 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 0.6 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.2 | 2.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 1.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 1.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 1.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.6 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.7 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 2.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 1.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.6 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 1.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.4 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 3.2 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.7 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.7 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 1.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.9 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.3 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 2.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.4 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 1.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 2.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.4 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 1.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 2.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 1.9 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 1.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.4 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) aromatase activity(GO:0070330) |
| 0.0 | 1.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.0 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 1.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.1 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 1.4 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 13.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.9 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 4.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 2.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 4.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 2.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 2.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 1.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 2.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 4.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 4.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 0.9 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 2.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 1.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 3.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 2.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 0.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 1.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.7 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.0 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.4 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 1.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 2.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |