Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Hsf4
Z-value: 1.45
Transcription factors associated with Hsf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsf4
|
ENSMUSG00000033249.4 | heat shock transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsf4 | mm10_v2_chr8_+_105269837_105269874 | 0.56 | 3.5e-04 | Click! |
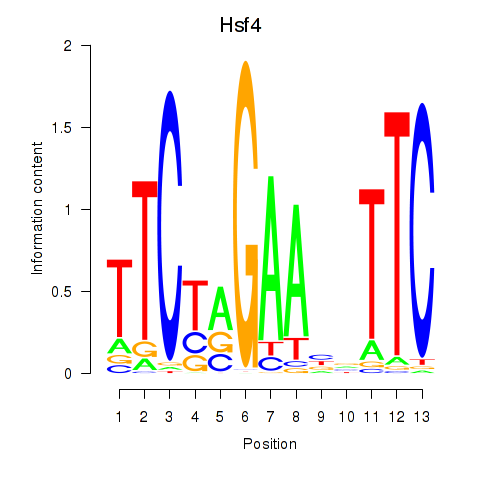
Activity profile of Hsf4 motif
Sorted Z-values of Hsf4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_8131982 | 8.48 |
ENSMUST00000065651.4
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr19_-_8405060 | 6.56 |
ENSMUST00000064507.5
ENSMUST00000120540.1 ENSMUST00000096269.4 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr6_-_138073196 | 4.02 |
ENSMUST00000050132.3
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr7_+_140845562 | 3.44 |
ENSMUST00000035300.5
|
Scgb1c1
|
secretoglobin, family 1C, member 1 |
| chr19_+_58670358 | 3.42 |
ENSMUST00000057270.7
|
Pnlip
|
pancreatic lipase |
| chr10_+_76562270 | 3.22 |
ENSMUST00000009259.4
ENSMUST00000105414.1 |
Spatc1l
|
spermatogenesis and centriole associated 1 like |
| chr6_-_128438673 | 3.00 |
ENSMUST00000032508.4
|
Fkbp4
|
FK506 binding protein 4 |
| chr12_-_110696289 | 2.99 |
ENSMUST00000021698.6
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr18_-_10706688 | 2.92 |
ENSMUST00000002549.7
ENSMUST00000117726.1 ENSMUST00000117828.1 |
Abhd3
|
abhydrolase domain containing 3 |
| chr1_+_195017399 | 2.91 |
ENSMUST00000181273.1
|
A330023F24Rik
|
RIKEN cDNA A330023F24 gene |
| chr17_+_35439155 | 2.86 |
ENSMUST00000071951.6
ENSMUST00000078205.7 ENSMUST00000116598.3 ENSMUST00000076256.7 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr3_-_107943390 | 2.80 |
ENSMUST00000106681.1
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr7_-_114562945 | 2.79 |
ENSMUST00000119712.1
ENSMUST00000032908.8 |
Cyp2r1
|
cytochrome P450, family 2, subfamily r, polypeptide 1 |
| chr5_-_149636164 | 2.75 |
ENSMUST00000076410.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr5_+_151368683 | 2.72 |
ENSMUST00000181114.1
ENSMUST00000181555.1 |
1700028E10Rik
|
RIKEN cDNA 1700028E10 gene |
| chr7_-_25539845 | 2.62 |
ENSMUST00000066503.7
ENSMUST00000064862.6 |
Ceacam2
|
carcinoembryonic antigen-related cell adhesion molecule 2 |
| chr3_-_107943362 | 2.56 |
ENSMUST00000106683.1
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr7_-_25539950 | 2.55 |
ENSMUST00000044547.8
|
Ceacam2
|
carcinoembryonic antigen-related cell adhesion molecule 2 |
| chrX_-_38635066 | 2.53 |
ENSMUST00000058265.7
|
C1galt1c1
|
C1GALT1-specific chaperone 1 |
| chr12_-_110695860 | 2.35 |
ENSMUST00000149189.1
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr4_+_40722461 | 2.34 |
ENSMUST00000030118.3
|
Dnaja1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr5_-_28055440 | 2.33 |
ENSMUST00000181503.1
|
Gm26608
|
predicted gene, 26608 |
| chr7_-_119523477 | 2.31 |
ENSMUST00000033267.2
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr11_-_106579111 | 2.26 |
ENSMUST00000103070.2
|
Tex2
|
testis expressed gene 2 |
| chr17_+_35470083 | 2.24 |
ENSMUST00000174525.1
ENSMUST00000068291.6 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr8_-_45382198 | 2.18 |
ENSMUST00000093526.6
|
Fam149a
|
family with sequence similarity 149, member A |
| chr12_-_110696248 | 2.15 |
ENSMUST00000124156.1
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr10_+_75893398 | 2.11 |
ENSMUST00000009236.4
|
Derl3
|
Der1-like domain family, member 3 |
| chr17_+_35424870 | 2.05 |
ENSMUST00000113879.3
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr5_+_92555035 | 2.02 |
ENSMUST00000146417.2
|
Fam47e
|
family with sequence similarity 47, member E |
| chr5_-_45450221 | 2.01 |
ENSMUST00000015950.5
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr5_-_45450143 | 2.00 |
ENSMUST00000154962.1
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr8_-_71537402 | 1.99 |
ENSMUST00000051672.7
|
Bst2
|
bone marrow stromal cell antigen 2 |
| chr19_+_3986564 | 1.98 |
ENSMUST00000054030.7
|
Acy3
|
aspartoacylase (aminoacylase) 3 |
| chr19_-_5802640 | 1.97 |
ENSMUST00000173523.1
ENSMUST00000173499.1 ENSMUST00000172812.2 |
Malat1
|
metastasis associated lung adenocarcinoma transcript 1 (non-coding RNA) |
| chr5_-_66151903 | 1.93 |
ENSMUST00000167950.1
|
Rbm47
|
RNA binding motif protein 47 |
| chr6_-_83677807 | 1.88 |
ENSMUST00000037882.6
|
Cd207
|
CD207 antigen |
| chr5_-_45450121 | 1.88 |
ENSMUST00000127562.1
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr5_+_135887905 | 1.81 |
ENSMUST00000005077.6
|
Hspb1
|
heat shock protein 1 |
| chr5_+_135887988 | 1.81 |
ENSMUST00000111155.1
|
Hspb1
|
heat shock protein 1 |
| chr17_+_35424842 | 1.80 |
ENSMUST00000174699.1
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr17_+_3326552 | 1.79 |
ENSMUST00000169838.1
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr7_-_24236632 | 1.76 |
ENSMUST00000037448.6
|
Zfp109
|
zinc finger protein 109 |
| chr10_+_29313500 | 1.76 |
ENSMUST00000020034.4
|
Echdc1
|
enoyl Coenzyme A hydratase domain containing 1 |
| chr10_-_86705485 | 1.73 |
ENSMUST00000020238.7
|
Hsp90b1
|
heat shock protein 90, beta (Grp94), member 1 |
| chr7_+_44198191 | 1.72 |
ENSMUST00000085450.2
|
Klk1b3
|
kallikrein 1-related peptidase b3 |
| chr15_+_62039216 | 1.71 |
ENSMUST00000183297.1
|
Pvt1
|
plasmacytoma variant translocation 1 |
| chr19_-_7802578 | 1.70 |
ENSMUST00000120522.1
ENSMUST00000065634.7 |
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr6_-_59024470 | 1.69 |
ENSMUST00000089860.5
|
Fam13a
|
family with sequence similarity 13, member A |
| chr5_-_151369172 | 1.64 |
ENSMUST00000067770.3
|
D730045B01Rik
|
RIKEN cDNA D730045B01 gene |
| chr13_-_23574196 | 1.64 |
ENSMUST00000105106.1
|
Hist1h2bf
|
histone cluster 1, H2bf |
| chr4_+_155601414 | 1.62 |
ENSMUST00000105608.2
|
Slc35e2
|
solute carrier family 35, member E2 |
| chr17_+_3532554 | 1.61 |
ENSMUST00000168560.1
|
Cldn20
|
claudin 20 |
| chrX_-_73716145 | 1.61 |
ENSMUST00000002091.5
|
Bcap31
|
B cell receptor associated protein 31 |
| chr1_+_88087802 | 1.58 |
ENSMUST00000113139.1
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr11_-_23497867 | 1.58 |
ENSMUST00000128559.1
ENSMUST00000147157.1 ENSMUST00000109539.1 |
Ahsa2
|
AHA1, activator of heat shock protein ATPase 2 |
| chr18_-_3337539 | 1.57 |
ENSMUST00000142690.1
ENSMUST00000025069.4 ENSMUST00000082141.5 ENSMUST00000165086.1 ENSMUST00000149803.1 |
Crem
|
cAMP responsive element modulator |
| chr18_-_3337614 | 1.57 |
ENSMUST00000150235.1
ENSMUST00000154470.1 |
Crem
|
cAMP responsive element modulator |
| chr6_+_3993776 | 1.55 |
ENSMUST00000031673.5
|
Gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr2_-_28466266 | 1.55 |
ENSMUST00000127683.1
ENSMUST00000086370.4 |
1700007K13Rik
|
RIKEN cDNA 1700007K13 gene |
| chr16_+_23107413 | 1.52 |
ENSMUST00000023599.6
ENSMUST00000168891.1 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr12_+_112106683 | 1.51 |
ENSMUST00000079400.4
|
Aspg
|
asparaginase homolog (S. cerevisiae) |
| chr8_+_109990430 | 1.50 |
ENSMUST00000001720.7
ENSMUST00000143741.1 |
Tat
|
tyrosine aminotransferase |
| chr9_-_105495130 | 1.49 |
ENSMUST00000038118.7
|
Atp2c1
|
ATPase, Ca++-sequestering |
| chr10_-_39163794 | 1.49 |
ENSMUST00000076713.4
|
Wisp3
|
WNT1 inducible signaling pathway protein 3 |
| chr3_+_114904062 | 1.49 |
ENSMUST00000081752.6
|
Olfm3
|
olfactomedin 3 |
| chr12_-_110696332 | 1.46 |
ENSMUST00000094361.4
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr7_+_30712209 | 1.45 |
ENSMUST00000005692.6
ENSMUST00000170371.1 |
Atp4a
|
ATPase, H+/K+ exchanging, gastric, alpha polypeptide |
| chr11_+_21239279 | 1.44 |
ENSMUST00000006221.7
ENSMUST00000109578.1 |
Vps54
|
vacuolar protein sorting 54 (yeast) |
| chr2_+_116900152 | 1.43 |
ENSMUST00000126467.1
ENSMUST00000128305.1 ENSMUST00000155323.1 |
D330050G23Rik
|
RIKEN cDNA D330050G23 gene |
| chr1_+_88095054 | 1.42 |
ENSMUST00000150634.1
ENSMUST00000058237.7 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr19_-_44069526 | 1.40 |
ENSMUST00000170801.1
|
Erlin1
|
ER lipid raft associated 1 |
| chrX_-_74023908 | 1.39 |
ENSMUST00000033769.8
ENSMUST00000114352.1 ENSMUST00000068286.5 ENSMUST00000114360.3 ENSMUST00000114354.3 |
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr16_+_23107754 | 1.37 |
ENSMUST00000077605.5
ENSMUST00000115341.3 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr17_-_12675833 | 1.37 |
ENSMUST00000024596.8
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr17_+_35262730 | 1.37 |
ENSMUST00000172785.1
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr7_-_46715676 | 1.35 |
ENSMUST00000006956.7
|
Saa3
|
serum amyloid A 3 |
| chr15_+_33083110 | 1.33 |
ENSMUST00000042167.9
|
Cpq
|
carboxypeptidase Q |
| chr17_-_45592569 | 1.31 |
ENSMUST00000163492.1
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chrX_-_74023745 | 1.30 |
ENSMUST00000114353.3
ENSMUST00000101458.2 |
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr17_-_12916345 | 1.30 |
ENSMUST00000079121.3
|
Mrpl18
|
mitochondrial ribosomal protein L18 |
| chr19_-_29812952 | 1.29 |
ENSMUST00000099525.3
|
Ranbp6
|
RAN binding protein 6 |
| chr2_+_172472512 | 1.28 |
ENSMUST00000029007.2
|
Fam209
|
family with sequence similarity 209 |
| chr19_+_47854970 | 1.28 |
ENSMUST00000026050.7
|
Gsto1
|
glutathione S-transferase omega 1 |
| chr6_+_125049952 | 1.27 |
ENSMUST00000088294.5
ENSMUST00000032481.7 |
Acrbp
|
proacrosin binding protein |
| chr8_-_22398588 | 1.27 |
ENSMUST00000033871.6
|
Slc25a15
|
solute carrier family 25 (mitochondrial carrier ornithine transporter), member 15 |
| chr11_-_26210553 | 1.26 |
ENSMUST00000101447.3
|
5730522E02Rik
|
RIKEN cDNA 5730522E02 gene |
| chr13_-_111490111 | 1.25 |
ENSMUST00000047627.7
|
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr6_-_59024340 | 1.24 |
ENSMUST00000173193.1
|
Fam13a
|
family with sequence similarity 13, member A |
| chr19_-_44069690 | 1.23 |
ENSMUST00000169092.1
|
Erlin1
|
ER lipid raft associated 1 |
| chr16_-_88751628 | 1.23 |
ENSMUST00000053149.2
|
Krtap13
|
keratin associated protein 13 |
| chr4_-_40722307 | 1.22 |
ENSMUST00000181475.1
|
Gm6297
|
predicted gene 6297 |
| chrX_-_109013389 | 1.22 |
ENSMUST00000033597.8
|
Hmgn5
|
high-mobility group nucleosome binding domain 5 |
| chr15_-_81399594 | 1.19 |
ENSMUST00000023039.8
|
St13
|
suppression of tumorigenicity 13 |
| chr9_-_105495037 | 1.17 |
ENSMUST00000176190.1
ENSMUST00000163879.2 ENSMUST00000112558.2 ENSMUST00000176390.1 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr2_-_175133322 | 1.16 |
ENSMUST00000099029.3
|
Gm14399
|
predicted gene 14399 |
| chr6_-_124888192 | 1.16 |
ENSMUST00000024044.6
|
Cd4
|
CD4 antigen |
| chr3_+_79591356 | 1.15 |
ENSMUST00000029382.7
|
Ppid
|
peptidylprolyl isomerase D (cyclophilin D) |
| chr5_+_92331828 | 1.14 |
ENSMUST00000125462.1
ENSMUST00000121096.1 ENSMUST00000113083.2 |
Art3
|
ADP-ribosyltransferase 3 |
| chr5_-_123749393 | 1.12 |
ENSMUST00000057795.5
ENSMUST00000111515.1 ENSMUST00000182309.1 |
Rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr13_-_111490028 | 1.12 |
ENSMUST00000091236.4
|
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr8_-_120177440 | 1.12 |
ENSMUST00000048786.6
|
Fam92b
|
family with sequence similarity 92, member B |
| chr4_+_129287614 | 1.10 |
ENSMUST00000102599.3
|
Sync
|
syncoilin |
| chr4_-_135385645 | 1.07 |
ENSMUST00000105857.1
ENSMUST00000105858.1 ENSMUST00000064481.8 ENSMUST00000123632.1 |
Ncmap
|
noncompact myelin associated protein |
| chr3_+_30792876 | 1.07 |
ENSMUST00000029256.7
|
Sec62
|
SEC62 homolog (S. cerevisiae) |
| chr7_+_119895836 | 1.07 |
ENSMUST00000106518.1
ENSMUST00000054440.3 |
Lyrm1
|
LYR motif containing 1 |
| chr11_-_12412136 | 1.07 |
ENSMUST00000174874.1
|
Cobl
|
cordon-bleu WH2 repeat |
| chr5_-_149636331 | 1.06 |
ENSMUST00000074846.7
ENSMUST00000110498.1 ENSMUST00000127977.1 ENSMUST00000132412.1 |
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr5_+_92331818 | 1.05 |
ENSMUST00000154245.1
|
Art3
|
ADP-ribosyltransferase 3 |
| chr13_-_51203065 | 1.05 |
ENSMUST00000091708.4
|
Hist1h2al
|
histone cluster 1, H2al |
| chr8_-_84893887 | 1.05 |
ENSMUST00000003907.7
ENSMUST00000182458.1 ENSMUST00000109745.1 ENSMUST00000142748.1 |
Gcdh
|
glutaryl-Coenzyme A dehydrogenase |
| chr17_+_21491256 | 1.04 |
ENSMUST00000076664.6
|
Zfp53
|
zinc finger protein 53 |
| chr4_+_155601854 | 1.03 |
ENSMUST00000118607.1
|
Slc35e2
|
solute carrier family 35, member E2 |
| chr13_-_48273865 | 1.03 |
ENSMUST00000180777.1
|
A330048O09Rik
|
RIKEN cDNA A330048O09 gene |
| chr15_-_3995708 | 1.03 |
ENSMUST00000046633.8
|
AW549877
|
expressed sequence AW549877 |
| chr9_-_105521147 | 1.02 |
ENSMUST00000176770.1
ENSMUST00000085133.6 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr17_-_56121946 | 1.02 |
ENSMUST00000041357.7
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr9_+_120149733 | 1.02 |
ENSMUST00000068698.7
ENSMUST00000093773.1 ENSMUST00000111627.1 |
Mobp
|
myelin-associated oligodendrocytic basic protein |
| chr5_+_30281377 | 1.02 |
ENSMUST00000101448.3
|
Drc1
|
dynein regulatory complex subunit 1 |
| chr9_+_75410145 | 1.00 |
ENSMUST00000180533.1
ENSMUST00000180574.1 |
4933433G15Rik
|
RIKEN cDNA 4933433G15 gene |
| chr4_-_96507386 | 1.00 |
ENSMUST00000124729.3
|
Cyp2j8
|
cytochrome P450, family 2, subfamily j, polypeptide 8 |
| chr9_+_18292267 | 0.99 |
ENSMUST00000001825.7
|
Chordc1
|
cysteine and histidine-rich domain (CHORD)-containing, zinc-binding protein 1 |
| chr7_+_43995833 | 0.98 |
ENSMUST00000007156.4
|
Klk1b11
|
kallikrein 1-related peptidase b11 |
| chr10_+_81268172 | 0.98 |
ENSMUST00000057798.8
|
Apba3
|
amyloid beta (A4) precursor protein-binding, family A, member 3 |
| chr9_-_109162021 | 0.97 |
ENSMUST00000120329.1
ENSMUST00000054925.6 |
Fbxw21
|
F-box and WD-40 domain protein 21 |
| chr4_+_148140699 | 0.97 |
ENSMUST00000140049.1
ENSMUST00000105707.1 |
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr10_-_86011833 | 0.96 |
ENSMUST00000105304.1
ENSMUST00000061699.5 |
Bpifc
|
BPI fold containing family C |
| chr8_-_3748941 | 0.95 |
ENSMUST00000012847.1
|
Cd209a
|
CD209a antigen |
| chr5_-_123749371 | 0.95 |
ENSMUST00000182955.1
ENSMUST00000182489.1 ENSMUST00000050827.7 |
Rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr11_+_21091291 | 0.94 |
ENSMUST00000093290.5
|
Peli1
|
pellino 1 |
| chrX_+_21714896 | 0.93 |
ENSMUST00000033414.7
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr15_-_81400043 | 0.92 |
ENSMUST00000172107.1
ENSMUST00000169204.1 ENSMUST00000163382.1 |
St13
|
suppression of tumorigenicity 13 |
| chr5_-_137962955 | 0.92 |
ENSMUST00000077119.6
|
Gjc3
|
gap junction protein, gamma 3 |
| chr4_+_40722912 | 0.91 |
ENSMUST00000164233.1
ENSMUST00000137246.1 ENSMUST00000125442.1 |
Dnaja1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr13_+_81657732 | 0.90 |
ENSMUST00000049055.6
|
Lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr15_-_82690499 | 0.90 |
ENSMUST00000100380.3
|
Cyp2d37-ps
|
cytochrome P450, family 2, subfamily d, polypeptide 37, pseudogene |
| chr12_-_79172609 | 0.90 |
ENSMUST00000055262.6
|
Vti1b
|
vesicle transport through interaction with t-SNAREs 1B |
| chr5_-_50058908 | 0.90 |
ENSMUST00000030971.5
|
Gpr125
|
G protein-coupled receptor 125 |
| chr2_-_110314525 | 0.88 |
ENSMUST00000133608.1
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
| chr6_+_51470339 | 0.88 |
ENSMUST00000094623.3
|
Cbx3
|
chromobox 3 |
| chr9_-_99436749 | 0.86 |
ENSMUST00000122384.1
|
Mras
|
muscle and microspikes RAS |
| chr2_-_37359235 | 0.86 |
ENSMUST00000112940.1
|
Pdcl
|
phosducin-like |
| chr5_-_116288944 | 0.85 |
ENSMUST00000086483.3
|
Ccdc60
|
coiled-coil domain containing 60 |
| chr17_-_56874421 | 0.85 |
ENSMUST00000043062.4
|
Acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr9_-_75409951 | 0.85 |
ENSMUST00000049355.10
|
Mapk6
|
mitogen-activated protein kinase 6 |
| chr11_+_52232183 | 0.85 |
ENSMUST00000109072.1
|
Skp1a
|
S-phase kinase-associated protein 1A |
| chr7_+_92561141 | 0.83 |
ENSMUST00000032842.6
ENSMUST00000085017.4 |
Ccdc90b
|
coiled-coil domain containing 90B |
| chr1_+_153874335 | 0.83 |
ENSMUST00000055314.3
|
Gm5531
|
predicted gene 5531 |
| chr11_-_93968242 | 0.82 |
ENSMUST00000107844.2
|
Nme1
|
NME/NM23 nucleoside diphosphate kinase 1 |
| chr4_+_155562348 | 0.82 |
ENSMUST00000030939.7
|
Nadk
|
NAD kinase |
| chr10_+_28668360 | 0.82 |
ENSMUST00000060409.6
ENSMUST00000056097.4 ENSMUST00000105516.2 |
Themis
|
thymocyte selection associated |
| chr2_-_37359274 | 0.82 |
ENSMUST00000009174.8
|
Pdcl
|
phosducin-like |
| chr11_+_52232009 | 0.82 |
ENSMUST00000037324.5
ENSMUST00000166537.1 |
Skp1a
|
S-phase kinase-associated protein 1A |
| chr19_+_26748268 | 0.82 |
ENSMUST00000175791.1
ENSMUST00000176698.1 ENSMUST00000177252.1 ENSMUST00000176475.1 ENSMUST00000112637.2 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr17_-_45595502 | 0.82 |
ENSMUST00000171081.1
ENSMUST00000172301.1 ENSMUST00000167332.1 ENSMUST00000170488.1 ENSMUST00000167195.1 ENSMUST00000064889.6 ENSMUST00000051574.6 ENSMUST00000164217.1 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr13_+_24831661 | 0.82 |
ENSMUST00000038039.2
|
Tdp2
|
tyrosyl-DNA phosphodiesterase 2 |
| chr2_-_103372725 | 0.81 |
ENSMUST00000139065.1
|
A930006I01Rik
|
RIKEN cDNA A930006I01 gene |
| chrX_-_61185558 | 0.81 |
ENSMUST00000166381.1
|
Cdr1
|
cerebellar degeneration related antigen 1 |
| chr8_-_64849818 | 0.81 |
ENSMUST00000034017.7
|
Klhl2
|
kelch-like 2, Mayven |
| chr5_+_29735940 | 0.80 |
ENSMUST00000114839.1
|
Dnajb6
|
DnaJ (Hsp40) homolog, subfamily B, member 6 |
| chr7_+_24257644 | 0.80 |
ENSMUST00000072713.6
|
Zfp108
|
zinc finger protein 108 |
| chr15_-_82354280 | 0.80 |
ENSMUST00000023085.5
|
Ndufa6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 (B14) |
| chr10_-_100589205 | 0.80 |
ENSMUST00000054471.8
|
4930430F08Rik
|
RIKEN cDNA 4930430F08 gene |
| chr3_+_81036360 | 0.79 |
ENSMUST00000029652.3
|
Pdgfc
|
platelet-derived growth factor, C polypeptide |
| chr7_-_70366735 | 0.77 |
ENSMUST00000089565.5
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr12_-_103631404 | 0.77 |
ENSMUST00000121625.1
ENSMUST00000044231.5 |
Serpina10
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr2_+_121456963 | 0.77 |
ENSMUST00000126764.1
|
Hypk
|
huntingtin interacting protein K |
| chr13_+_41655697 | 0.77 |
ENSMUST00000067176.8
|
Gm5082
|
predicted gene 5082 |
| chr7_+_136894598 | 0.75 |
ENSMUST00000081510.2
|
Mgmt
|
O-6-methylguanine-DNA methyltransferase |
| chr16_-_84835557 | 0.75 |
ENSMUST00000138279.1
ENSMUST00000023608.7 |
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr1_+_164249233 | 0.75 |
ENSMUST00000169394.1
|
Slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr1_+_91250482 | 0.75 |
ENSMUST00000171112.1
|
Ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr4_-_88683912 | 0.74 |
ENSMUST00000105147.1
|
Ifna2
|
interferon alpha 2 |
| chr5_-_139814025 | 0.74 |
ENSMUST00000146780.1
|
Tmem184a
|
transmembrane protein 184a |
| chr14_-_79390666 | 0.73 |
ENSMUST00000022597.7
|
Naa16
|
N(alpha)-acetyltransferase 16, NatA auxiliary subunit |
| chr7_-_126584578 | 0.73 |
ENSMUST00000150311.1
|
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr4_-_63861326 | 0.73 |
ENSMUST00000030047.2
|
Tnfsf8
|
tumor necrosis factor (ligand) superfamily, member 8 |
| chr9_+_119402444 | 0.73 |
ENSMUST00000035093.8
ENSMUST00000165044.1 |
Acvr2b
|
activin receptor IIB |
| chr17_+_53479212 | 0.73 |
ENSMUST00000017975.5
|
Rab5a
|
RAB5A, member RAS oncogene family |
| chr7_-_130519465 | 0.72 |
ENSMUST00000035458.7
ENSMUST00000033139.7 |
Ate1
|
arginyltransferase 1 |
| chr2_-_37359196 | 0.72 |
ENSMUST00000147703.1
|
Pdcl
|
phosducin-like |
| chr14_-_37135126 | 0.71 |
ENSMUST00000042564.9
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr16_+_88777829 | 0.71 |
ENSMUST00000009191.3
|
Gm5965
|
predicted gene 5965 |
| chr15_+_87625214 | 0.71 |
ENSMUST00000068088.6
|
Fam19a5
|
family with sequence similarity 19, member A5 |
| chr5_-_139814231 | 0.71 |
ENSMUST00000044002.4
|
Tmem184a
|
transmembrane protein 184a |
| chr10_+_127759721 | 0.70 |
ENSMUST00000073639.5
|
Rdh1
|
retinol dehydrogenase 1 (all trans) |
| chr5_+_29735991 | 0.70 |
ENSMUST00000012734.5
|
Dnajb6
|
DnaJ (Hsp40) homolog, subfamily B, member 6 |
| chr6_+_29853746 | 0.70 |
ENSMUST00000064872.6
ENSMUST00000152581.1 ENSMUST00000176265.1 ENSMUST00000154079.1 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr7_+_43975963 | 0.69 |
ENSMUST00000081399.3
|
Klk1b9
|
kallikrein 1-related peptidase b9 |
| chr6_+_48647224 | 0.69 |
ENSMUST00000078223.3
|
Gimap8
|
GTPase, IMAP family member 8 |
| chr5_-_92675253 | 0.69 |
ENSMUST00000151180.1
ENSMUST00000150359.1 |
Ccdc158
|
coiled-coil domain containing 158 |
| chr6_+_88084473 | 0.68 |
ENSMUST00000032143.6
|
Rpn1
|
ribophorin I |
| chr15_-_99528017 | 0.68 |
ENSMUST00000023750.7
|
Faim2
|
Fas apoptotic inhibitory molecule 2 |
| chr11_-_54860564 | 0.67 |
ENSMUST00000144164.1
|
Lyrm7
|
LYR motif containing 7 |
| chrX_+_161717055 | 0.66 |
ENSMUST00000112338.1
|
Rai2
|
retinoic acid induced 2 |
| chr5_-_88676135 | 0.66 |
ENSMUST00000078945.5
|
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr11_+_121146143 | 0.66 |
ENSMUST00000039088.8
ENSMUST00000155694.1 |
Tex19.1
|
testis expressed gene 19.1 |
| chr19_-_7039987 | 0.65 |
ENSMUST00000025918.7
|
Stip1
|
stress-induced phosphoprotein 1 |
| chr6_-_51469836 | 0.65 |
ENSMUST00000090002.7
|
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr9_-_110989611 | 0.65 |
ENSMUST00000084922.5
|
Rtp3
|
receptor transporter protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsf4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0070342 | brown fat cell proliferation(GO:0070342) regulation of brown fat cell proliferation(GO:0070347) |
| 1.5 | 8.9 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 1.1 | 15.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.9 | 3.7 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.8 | 2.4 | GO:1902605 | heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.7 | 5.9 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.5 | 3.8 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.5 | 1.6 | GO:1904154 | protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.5 | 1.4 | GO:0009804 | coumarin metabolic process(GO:0009804) phenylpropanoid catabolic process(GO:0046271) |
| 0.4 | 1.3 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.4 | 2.1 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.4 | 1.3 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.4 | 3.6 | GO:0099640 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) axo-dendritic protein transport(GO:0099640) |
| 0.4 | 2.0 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.4 | 2.0 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.4 | 1.2 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.4 | 2.7 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.4 | 1.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.4 | 1.5 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.4 | 2.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.4 | 3.3 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.4 | 3.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.3 | 1.0 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.3 | 1.3 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.3 | 1.3 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.3 | 0.9 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.3 | 2.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.3 | 1.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.3 | 1.4 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.3 | 1.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.3 | 0.8 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.2 | 1.0 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 | 0.7 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.2 | 1.4 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.2 | 0.7 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 0.9 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 0.7 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.2 | 1.5 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.2 | 0.6 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.2 | 8.8 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.2 | 0.6 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.2 | 3.4 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.2 | 0.4 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.2 | 1.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 0.6 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.2 | 0.5 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.2 | 1.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.2 | 0.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 0.6 | GO:1903984 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.2 | 0.8 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.2 | 0.8 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 0.6 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.7 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 1.0 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 1.0 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 1.6 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 3.0 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.4 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 0.4 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.1 | 0.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.4 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.9 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.1 | 0.6 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.5 | GO:1900108 | inner medullary collecting duct development(GO:0072061) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 1.0 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 1.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 1.7 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.4 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 0.8 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 2.6 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.7 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.3 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 0.8 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 1.4 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.3 | GO:0015755 | fructose transport(GO:0015755) |
| 0.1 | 0.4 | GO:0035962 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.1 | 0.8 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.9 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.1 | 0.4 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.1 | 0.4 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.7 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.7 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.3 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 0.3 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 3.5 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 0.6 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 1.3 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 0.8 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 1.9 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 1.4 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 0.3 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.7 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 1.4 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.1 | 0.4 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 1.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.7 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.9 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 3.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 0.5 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.1 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.1 | 0.7 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 1.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.5 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.2 | GO:2000812 | response to rapamycin(GO:1901355) regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 | 0.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 0.5 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.3 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 2.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 1.4 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 3.1 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 0.3 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.2 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.6 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.4 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.2 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.3 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.0 | 1.4 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.1 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 2.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.3 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 1.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.6 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.6 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.7 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.5 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:0009407 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.0 | 0.5 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 2.8 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.7 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.2 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.7 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.7 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0033483 | oxygen homeostasis(GO:0032364) gas homeostasis(GO:0033483) |
| 0.0 | 0.1 | GO:1902523 | negative regulation of interleukin-18 production(GO:0032701) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 1.1 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.2 | GO:0044273 | sulfur compound catabolic process(GO:0044273) |
| 0.0 | 0.1 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 1.0 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.6 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 1.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.3 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.4 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.4 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 1.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.2 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.1 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.5 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 8.9 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.9 | 10.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 1.6 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.3 | 2.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.2 | 3.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 0.7 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.2 | 1.4 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 1.7 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.2 | 5.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 1.0 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 2.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.5 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.9 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 1.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.0 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.6 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 0.6 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.2 | GO:0060473 | cortical granule(GO:0060473) |
| 0.1 | 1.1 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.8 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.9 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 2.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.5 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 1.0 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.9 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.5 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 5.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 2.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 2.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 3.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 5.2 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:1990712 | dense body(GO:0097433) HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.6 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 1.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 1.3 | 7.7 | GO:0002135 | CTP binding(GO:0002135) |
| 1.0 | 15.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 1.0 | 2.9 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.9 | 3.7 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.8 | 10.6 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.7 | 2.1 | GO:0032564 | dATP binding(GO:0032564) |
| 0.6 | 3.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.6 | 1.8 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.5 | 1.5 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.5 | 1.4 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.5 | 1.4 | GO:0050610 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.5 | 1.4 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.4 | 3.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.4 | 3.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 1.0 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.3 | 1.3 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.3 | 0.9 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.3 | 2.4 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.3 | 1.2 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.3 | 2.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.3 | 1.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.2 | 5.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 1.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.2 | 2.5 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.2 | 1.7 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 0.6 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 3.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 0.8 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.2 | 0.9 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.2 | 2.0 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 1.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.2 | 2.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 1.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 1.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 0.6 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 2.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 5.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 2.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 0.5 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.2 | 1.5 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.1 | 0.7 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 0.6 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.4 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 0.8 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 1.0 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.5 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 5.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 2.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 2.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.7 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 3.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.7 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.6 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 1.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.9 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.3 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.3 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 1.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.9 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 1.9 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.9 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.2 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.1 | 3.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.3 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.1 | 0.7 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.0 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 1.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 2.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 4.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.9 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 1.8 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.6 | GO:0043176 | amine binding(GO:0043176) |
| 0.0 | 1.0 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.7 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.7 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 1.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 1.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 3.7 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 3.8 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.7 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.6 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.7 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 4.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 3.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.8 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 3.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.6 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 2.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.4 | 8.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 1.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.3 | 3.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 1.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 2.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 5.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 1.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 11.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 3.4 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 3.7 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.1 | 0.6 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 2.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 2.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.0 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 2.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.9 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |