Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Hoxd11_Cdx1_Hoxc11
Z-value: 2.27
Transcription factors associated with Hoxd11_Cdx1_Hoxc11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
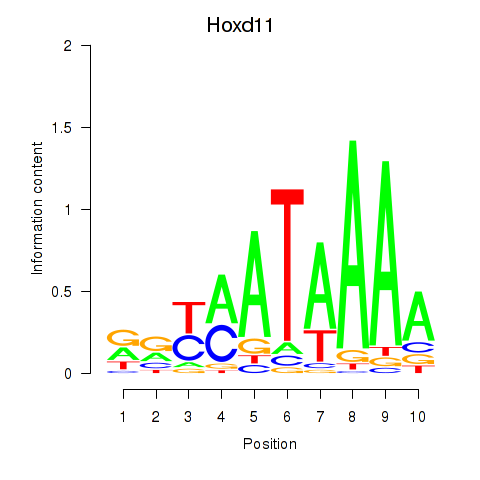
Hoxd11
|
ENSMUSG00000042499.12 | homeobox D11 |
|
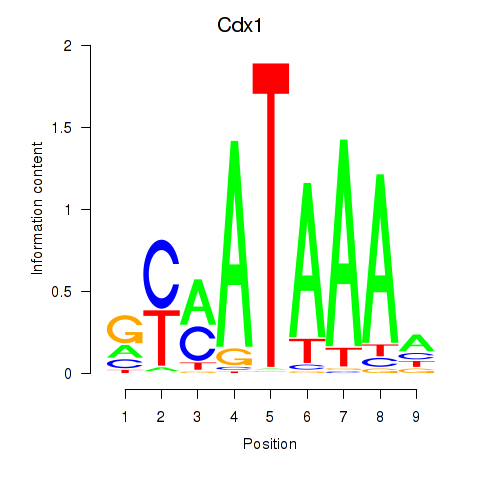
Cdx1
|
ENSMUSG00000024619.8 | caudal type homeobox 1 |
|
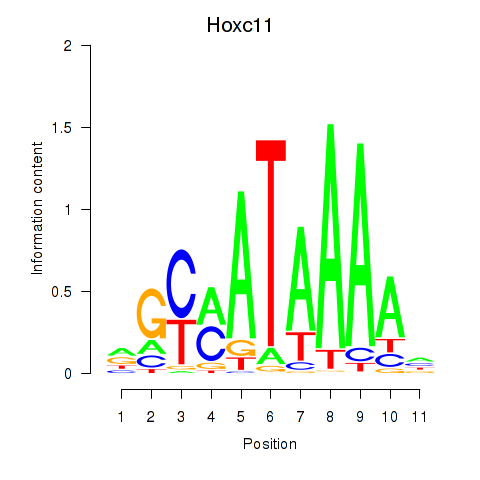
Hoxc11
|
ENSMUSG00000001656.3 | homeobox C11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxc11 | mm10_v2_chr15_+_102954427_102954427 | 0.56 | 4.1e-04 | Click! |
| Hoxd11 | mm10_v2_chr2_+_74681991_74682007 | 0.51 | 1.6e-03 | Click! |
| Cdx1 | mm10_v2_chr18_-_61036189_61036210 | 0.35 | 3.7e-02 | Click! |
Activity profile of Hoxd11_Cdx1_Hoxc11 motif
Sorted Z-values of Hoxd11_Cdx1_Hoxc11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_142661858 | 26.03 |
ENSMUST00000145896.2
|
Igf2
|
insulin-like growth factor 2 |
| chr2_+_84988194 | 14.26 |
ENSMUST00000028466.5
|
Prg3
|
proteoglycan 3 |
| chr14_-_37110087 | 9.22 |
ENSMUST00000179488.1
|
2610528A11Rik
|
RIKEN cDNA 2610528A11 gene |
| chrX_+_96455359 | 8.47 |
ENSMUST00000033553.7
|
Heph
|
hephaestin |
| chr1_+_107535508 | 7.94 |
ENSMUST00000182198.1
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr19_-_45812291 | 6.77 |
ENSMUST00000086993.4
|
Kcnip2
|
Kv channel-interacting protein 2 |
| chr17_+_5799491 | 6.44 |
ENSMUST00000181484.1
|
3300005D01Rik
|
RIKEN cDNA 3300005D01 gene |
| chr1_-_45503282 | 6.34 |
ENSMUST00000086430.4
|
Col5a2
|
collagen, type V, alpha 2 |
| chr2_+_155611175 | 5.90 |
ENSMUST00000092995.5
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr9_+_62858085 | 5.64 |
ENSMUST00000034777.6
ENSMUST00000163820.1 |
Calml4
|
calmodulin-like 4 |
| chrX_+_106920618 | 5.32 |
ENSMUST00000060576.7
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr1_-_89933290 | 5.32 |
ENSMUST00000036954.7
|
Gbx2
|
gastrulation brain homeobox 2 |
| chr10_+_119992916 | 5.25 |
ENSMUST00000105261.2
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr10_+_45577811 | 5.17 |
ENSMUST00000037044.6
|
Hace1
|
HECT domain and ankyrin repeat containing, E3 ubiquitin protein ligase 1 |
| chr17_-_43502773 | 5.09 |
ENSMUST00000024707.8
ENSMUST00000117137.1 |
Mep1a
|
meprin 1 alpha |
| chr17_+_71019548 | 5.07 |
ENSMUST00000073211.5
ENSMUST00000179759.1 |
Myom1
|
myomesin 1 |
| chr9_+_65890237 | 4.72 |
ENSMUST00000045802.6
|
2810417H13Rik
|
RIKEN cDNA 2810417H13 gene |
| chr17_+_71019503 | 4.70 |
ENSMUST00000024847.7
|
Myom1
|
myomesin 1 |
| chr4_-_133968611 | 4.43 |
ENSMUST00000102552.1
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr3_-_123034943 | 4.43 |
ENSMUST00000029761.7
|
Myoz2
|
myozenin 2 |
| chr15_+_81936753 | 4.38 |
ENSMUST00000038757.7
|
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr15_-_78855517 | 4.38 |
ENSMUST00000044584.4
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr5_-_73191848 | 4.35 |
ENSMUST00000176910.1
|
Fryl
|
furry homolog-like (Drosophila) |
| chr19_+_53529100 | 4.31 |
ENSMUST00000038287.6
|
Dusp5
|
dual specificity phosphatase 5 |
| chr4_+_11758147 | 4.22 |
ENSMUST00000029871.5
ENSMUST00000108303.1 |
Cdh17
|
cadherin 17 |
| chr14_+_74640840 | 4.09 |
ENSMUST00000036653.3
|
Htr2a
|
5-hydroxytryptamine (serotonin) receptor 2A |
| chr1_-_163289214 | 4.08 |
ENSMUST00000183691.1
|
Prrx1
|
paired related homeobox 1 |
| chrX_+_164140447 | 4.07 |
ENSMUST00000073973.4
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr4_-_154636831 | 3.97 |
ENSMUST00000030902.6
ENSMUST00000105637.1 ENSMUST00000070313.7 ENSMUST00000105636.1 ENSMUST00000105638.2 ENSMUST00000097759.2 ENSMUST00000124771.1 |
Prdm16
|
PR domain containing 16 |
| chr11_-_99244058 | 3.93 |
ENSMUST00000103132.3
ENSMUST00000038214.6 |
Krt222
|
keratin 222 |
| chr1_+_63176818 | 3.92 |
ENSMUST00000129339.1
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr7_-_134232005 | 3.89 |
ENSMUST00000134504.1
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr15_+_81936911 | 3.85 |
ENSMUST00000135663.1
|
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr13_-_84064772 | 3.75 |
ENSMUST00000182477.1
|
Gm17750
|
predicted gene, 17750 |
| chr6_-_52226165 | 3.62 |
ENSMUST00000114425.2
|
Hoxa9
|
homeobox A9 |
| chr17_+_5799616 | 3.60 |
ENSMUST00000181392.1
|
3300005D01Rik
|
RIKEN cDNA 3300005D01 gene |
| chr7_-_134232125 | 3.45 |
ENSMUST00000127524.1
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr6_-_139501907 | 3.41 |
ENSMUST00000170650.1
|
Rergl
|
RERG/RAS-like |
| chr5_-_73256555 | 3.33 |
ENSMUST00000171179.1
ENSMUST00000101127.5 |
Fryl
Fryl
|
furry homolog-like (Drosophila) furry homolog-like (Drosophila) |
| chr19_-_7341792 | 3.21 |
ENSMUST00000164205.1
ENSMUST00000165286.1 ENSMUST00000168324.1 ENSMUST00000032557.8 |
Mark2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr8_+_69902178 | 3.19 |
ENSMUST00000050373.5
|
Tssk6
|
testis-specific serine kinase 6 |
| chr4_-_133967296 | 3.15 |
ENSMUST00000105893.1
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr3_-_144760841 | 3.13 |
ENSMUST00000059091.5
|
Clca1
|
chloride channel calcium activated 1 |
| chr14_+_65805832 | 3.08 |
ENSMUST00000022612.3
|
Pbk
|
PDZ binding kinase |
| chr2_-_32387760 | 3.07 |
ENSMUST00000050785.8
|
Lcn2
|
lipocalin 2 |
| chr8_+_57332111 | 3.07 |
ENSMUST00000181638.1
|
5033428I22Rik
|
RIKEN cDNA 5033428I22 gene |
| chr14_+_65806066 | 3.05 |
ENSMUST00000139644.1
|
Pbk
|
PDZ binding kinase |
| chr9_-_39604124 | 3.01 |
ENSMUST00000042485.4
ENSMUST00000141370.1 |
AW551984
|
expressed sequence AW551984 |
| chr19_-_7341848 | 2.92 |
ENSMUST00000171393.1
|
Mark2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr16_+_26463135 | 2.90 |
ENSMUST00000161053.1
ENSMUST00000115302.1 |
Cldn16
|
claudin 16 |
| chr9_+_43310763 | 2.76 |
ENSMUST00000034511.5
|
Trim29
|
tripartite motif-containing 29 |
| chr3_-_144819494 | 2.75 |
ENSMUST00000029929.7
|
Clca2
|
chloride channel calcium activated 2 |
| chr11_+_96323253 | 2.73 |
ENSMUST00000093944.3
|
Hoxb3
|
homeobox B3 |
| chr9_-_42124276 | 2.72 |
ENSMUST00000060989.8
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing |
| chr14_+_34053466 | 2.71 |
ENSMUST00000058725.3
|
Antxrl
|
anthrax toxin receptor-like |
| chr5_-_24030649 | 2.70 |
ENSMUST00000030849.6
|
Fam126a
|
family with sequence similarity 126, member A |
| chr4_-_88033328 | 2.70 |
ENSMUST00000078090.5
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr18_-_32036941 | 2.70 |
ENSMUST00000134663.1
|
Myo7b
|
myosin VIIB |
| chr14_+_26894557 | 2.67 |
ENSMUST00000090337.4
ENSMUST00000165929.2 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr8_+_57455898 | 2.65 |
ENSMUST00000034023.3
|
Scrg1
|
scrapie responsive gene 1 |
| chr9_-_123678873 | 2.64 |
ENSMUST00000040960.6
|
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr15_+_85510812 | 2.60 |
ENSMUST00000079690.2
|
Gm4825
|
predicted pseudogene 4825 |
| chr4_-_136602641 | 2.51 |
ENSMUST00000105847.1
ENSMUST00000116273.2 |
Kdm1a
|
lysine (K)-specific demethylase 1A |
| chr3_+_122044428 | 2.45 |
ENSMUST00000013995.8
|
Abca4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr15_-_79062866 | 2.45 |
ENSMUST00000151889.1
ENSMUST00000040676.4 |
Ankrd54
|
ankyrin repeat domain 54 |
| chr9_-_123678782 | 2.45 |
ENSMUST00000170591.1
ENSMUST00000171647.1 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr19_+_60755947 | 2.43 |
ENSMUST00000088237.4
|
Nanos1
|
nanos homolog 1 (Drosophila) |
| chr7_+_25306085 | 2.42 |
ENSMUST00000119703.1
ENSMUST00000108409.1 |
Tmem145
|
transmembrane protein 145 |
| chr11_-_120549695 | 2.40 |
ENSMUST00000034913.4
|
Fam195b
|
family with sequence similarity 195, member B |
| chr10_-_13324160 | 2.35 |
ENSMUST00000105545.4
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr18_-_15063560 | 2.33 |
ENSMUST00000168989.1
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr18_+_21072329 | 2.30 |
ENSMUST00000082235.4
|
Mep1b
|
meprin 1 beta |
| chr7_+_66109474 | 2.28 |
ENSMUST00000036372.6
|
Chsy1
|
chondroitin sulfate synthase 1 |
| chrX_+_56447965 | 2.25 |
ENSMUST00000079663.6
|
Gm2174
|
predicted gene 2174 |
| chr16_-_76022266 | 2.23 |
ENSMUST00000114240.1
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr17_-_35074485 | 2.22 |
ENSMUST00000007259.3
|
Ly6g6d
|
lymphocyte antigen 6 complex, locus G6D |
| chr11_+_96316684 | 2.11 |
ENSMUST00000049241.7
|
Hoxb4
|
homeobox B4 |
| chr3_+_84666192 | 2.10 |
ENSMUST00000107682.1
|
Tmem154
|
transmembrane protein 154 |
| chr9_+_110763646 | 2.09 |
ENSMUST00000079784.7
|
Myl3
|
myosin, light polypeptide 3 |
| chr6_-_83536215 | 2.06 |
ENSMUST00000075161.5
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr10_+_37139558 | 2.04 |
ENSMUST00000062667.3
|
5930403N24Rik
|
RIKEN cDNA 5930403N24 gene |
| chr15_-_101491509 | 2.01 |
ENSMUST00000023718.7
|
5430421N21Rik
|
RIKEN cDNA 5430421N21 gene |
| chrX_+_153139941 | 1.97 |
ENSMUST00000039720.4
ENSMUST00000144175.2 |
Rragb
|
Ras-related GTP binding B |
| chr10_+_69787431 | 1.97 |
ENSMUST00000183240.1
|
Ank3
|
ankyrin 3, epithelial |
| chr7_-_135528645 | 1.96 |
ENSMUST00000053716.7
|
Clrn3
|
clarin 3 |
| chr16_+_10812915 | 1.93 |
ENSMUST00000115822.1
|
Gm11172
|
predicted gene 11172 |
| chr11_-_109472611 | 1.92 |
ENSMUST00000168740.1
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr11_-_17008647 | 1.91 |
ENSMUST00000102881.3
|
Plek
|
pleckstrin |
| chr4_+_5724304 | 1.86 |
ENSMUST00000108380.1
|
Fam110b
|
family with sequence similarity 110, member B |
| chr7_-_30612731 | 1.82 |
ENSMUST00000006476.4
|
Upk1a
|
uroplakin 1A |
| chr2_-_84822546 | 1.81 |
ENSMUST00000028471.5
|
Smtnl1
|
smoothelin-like 1 |
| chr7_+_131032061 | 1.79 |
ENSMUST00000084509.3
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr17_-_78906899 | 1.76 |
ENSMUST00000042683.6
ENSMUST00000169544.1 |
Sult6b1
|
sulfotransferase family, cytosolic, 6B, member 1 |
| chr16_+_37011758 | 1.76 |
ENSMUST00000071452.5
ENSMUST00000054034.6 |
Polq
|
polymerase (DNA directed), theta |
| chr6_-_87981482 | 1.74 |
ENSMUST00000056403.5
|
H1fx
|
H1 histone family, member X |
| chr17_-_70998010 | 1.73 |
ENSMUST00000024846.6
|
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr14_+_56042123 | 1.72 |
ENSMUST00000015576.4
|
Mcpt2
|
mast cell protease 2 |
| chr11_+_29463735 | 1.69 |
ENSMUST00000155854.1
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr6_-_52218686 | 1.69 |
ENSMUST00000134367.2
|
Hoxa7
|
homeobox A7 |
| chr6_-_12749193 | 1.68 |
ENSMUST00000046121.6
ENSMUST00000172356.1 |
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr6_-_13839916 | 1.68 |
ENSMUST00000060442.7
|
Gpr85
|
G protein-coupled receptor 85 |
| chr2_+_57997884 | 1.65 |
ENSMUST00000112616.1
ENSMUST00000166729.1 |
Galnt5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 |
| chr4_-_64046925 | 1.65 |
ENSMUST00000107377.3
|
Tnc
|
tenascin C |
| chr10_+_116177351 | 1.64 |
ENSMUST00000155606.1
ENSMUST00000128399.1 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr1_-_172027251 | 1.54 |
ENSMUST00000138714.1
|
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chr9_-_64172879 | 1.52 |
ENSMUST00000176299.1
ENSMUST00000130127.1 ENSMUST00000176794.1 ENSMUST00000177045.1 |
Zwilch
|
zwilch kinetochore protein |
| chr6_+_125552948 | 1.50 |
ENSMUST00000112254.1
ENSMUST00000112253.1 ENSMUST00000001995.7 |
Vwf
|
Von Willebrand factor homolog |
| chr10_-_30655859 | 1.49 |
ENSMUST00000092610.4
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr18_-_6490808 | 1.49 |
ENSMUST00000028100.6
ENSMUST00000050542.5 |
Epc1
|
enhancer of polycomb homolog 1 (Drosophila) |
| chr3_-_49757257 | 1.48 |
ENSMUST00000035931.7
|
Pcdh18
|
protocadherin 18 |
| chr2_-_170194033 | 1.47 |
ENSMUST00000180625.1
|
Gm17619
|
predicted gene, 17619 |
| chr6_-_122340499 | 1.46 |
ENSMUST00000160843.1
|
Phc1
|
polyhomeotic-like 1 (Drosophila) |
| chr19_+_33822908 | 1.44 |
ENSMUST00000042061.6
|
Gm5519
|
predicted pseudogene 5519 |
| chr6_-_122340525 | 1.43 |
ENSMUST00000112600.2
|
Phc1
|
polyhomeotic-like 1 (Drosophila) |
| chr17_-_51826562 | 1.42 |
ENSMUST00000024720.4
ENSMUST00000129667.1 ENSMUST00000156051.1 ENSMUST00000169480.1 ENSMUST00000148559.1 |
Satb1
|
special AT-rich sequence binding protein 1 |
| chr8_-_57653023 | 1.40 |
ENSMUST00000034021.5
|
Galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr17_+_80307396 | 1.38 |
ENSMUST00000068175.5
|
Arhgef33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr4_-_133967893 | 1.36 |
ENSMUST00000100472.3
ENSMUST00000136327.1 |
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr2_-_114013619 | 1.32 |
ENSMUST00000090275.4
|
Gjd2
|
gap junction protein, delta 2 |
| chr19_-_50678642 | 1.29 |
ENSMUST00000072685.6
ENSMUST00000164039.2 |
Sorcs1
|
VPS10 domain receptor protein SORCS 1 |
| chr8_+_25720054 | 1.29 |
ENSMUST00000068916.8
ENSMUST00000139836.1 |
Ppapdc1b
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr13_+_19214103 | 1.29 |
ENSMUST00000103558.1
|
Tcrg-C1
|
T cell receptor gamma, constant 1 |
| chr4_-_11981265 | 1.26 |
ENSMUST00000098260.2
|
Gm10604
|
predicted gene 10604 |
| chr8_+_121116163 | 1.26 |
ENSMUST00000054691.6
|
Foxc2
|
forkhead box C2 |
| chr6_-_24168083 | 1.26 |
ENSMUST00000031713.8
|
Slc13a1
|
solute carrier family 13 (sodium/sulfate symporters), member 1 |
| chrX_+_56779437 | 1.24 |
ENSMUST00000114773.3
|
Fhl1
|
four and a half LIM domains 1 |
| chr7_+_46861203 | 1.23 |
ENSMUST00000014545.4
|
Ldhc
|
lactate dehydrogenase C |
| chr18_+_69593361 | 1.23 |
ENSMUST00000114978.2
ENSMUST00000114977.1 |
Tcf4
|
transcription factor 4 |
| chr1_-_152625212 | 1.23 |
ENSMUST00000027760.7
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr18_-_62756275 | 1.21 |
ENSMUST00000067450.1
ENSMUST00000048109.5 |
2700046A07Rik
|
RIKEN cDNA 2700046A07 gene |
| chr16_-_56795823 | 1.19 |
ENSMUST00000023437.4
|
Gpr128
|
G protein-coupled receptor 128 |
| chr14_+_76504185 | 1.18 |
ENSMUST00000177207.1
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr1_-_128328311 | 1.15 |
ENSMUST00000073490.6
|
Lct
|
lactase |
| chr18_-_47333311 | 1.14 |
ENSMUST00000126684.1
ENSMUST00000156422.1 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr8_+_57320975 | 1.14 |
ENSMUST00000040104.3
|
Hand2
|
heart and neural crest derivatives expressed transcript 2 |
| chr14_+_20674311 | 1.12 |
ENSMUST00000048657.8
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr1_+_58646608 | 1.11 |
ENSMUST00000081455.4
|
Gm10068
|
predicted gene 10068 |
| chr3_-_89960562 | 1.11 |
ENSMUST00000069805.7
|
Atp8b2
|
ATPase, class I, type 8B, member 2 |
| chr1_-_128102412 | 1.10 |
ENSMUST00000112538.1
ENSMUST00000086614.5 |
Zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chrX_-_74246534 | 1.10 |
ENSMUST00000101454.2
ENSMUST00000033699.6 |
Flna
|
filamin, alpha |
| chr4_-_133967953 | 1.08 |
ENSMUST00000102553.4
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr1_-_175688353 | 1.07 |
ENSMUST00000104984.1
|
Chml
|
choroideremia-like |
| chr5_+_137641334 | 1.05 |
ENSMUST00000177466.1
ENSMUST00000166099.2 |
Sap25
|
sin3 associated polypeptide |
| chr13_-_58354862 | 1.04 |
ENSMUST00000043605.5
|
Kif27
|
kinesin family member 27 |
| chr15_-_34356421 | 1.03 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chrX_+_13632769 | 0.99 |
ENSMUST00000096492.3
|
Gpr34
|
G protein-coupled receptor 34 |
| chr11_-_19018956 | 0.99 |
ENSMUST00000068264.7
ENSMUST00000144988.1 ENSMUST00000185131.1 |
Meis1
|
Meis homeobox 1 |
| chr14_-_106101157 | 0.96 |
ENSMUST00000169588.1
|
Gm4775
|
predicted gene 4775 |
| chr1_-_37496095 | 0.95 |
ENSMUST00000148047.1
ENSMUST00000143636.1 |
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr11_-_19018714 | 0.92 |
ENSMUST00000177417.1
|
Meis1
|
Meis homeobox 1 |
| chr2_-_140671462 | 0.90 |
ENSMUST00000110057.2
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr15_-_42676967 | 0.90 |
ENSMUST00000022921.5
|
Angpt1
|
angiopoietin 1 |
| chr11_-_100105626 | 0.90 |
ENSMUST00000107416.2
|
Krt36
|
keratin 36 |
| chr2_+_36230426 | 0.86 |
ENSMUST00000062069.5
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr15_+_11000716 | 0.86 |
ENSMUST00000117100.1
ENSMUST00000022851.7 |
Slc45a2
|
solute carrier family 45, member 2 |
| chr17_+_35059035 | 0.85 |
ENSMUST00000007255.6
ENSMUST00000174493.1 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr1_-_172027269 | 0.84 |
ENSMUST00000027837.6
ENSMUST00000111264.1 |
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chr15_+_100761741 | 0.83 |
ENSMUST00000023776.6
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr9_-_21067093 | 0.83 |
ENSMUST00000115494.2
|
Zglp1
|
zinc finger, GATA-like protein 1 |
| chr6_-_145047725 | 0.83 |
ENSMUST00000123930.1
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr6_+_125215551 | 0.82 |
ENSMUST00000032487.7
ENSMUST00000100942.2 ENSMUST00000063588.8 |
Vamp1
|
vesicle-associated membrane protein 1 |
| chr8_+_15057646 | 0.81 |
ENSMUST00000033842.3
|
Myom2
|
myomesin 2 |
| chr1_-_176807124 | 0.79 |
ENSMUST00000057037.7
|
Cep170
|
centrosomal protein 170 |
| chr11_-_97700327 | 0.79 |
ENSMUST00000018681.7
|
Pcgf2
|
polycomb group ring finger 2 |
| chrY_-_79667079 | 0.78 |
ENSMUST00000178903.1
|
Gm21242
|
predicted gene, 21242 |
| chr3_-_63851251 | 0.77 |
ENSMUST00000162269.2
ENSMUST00000159676.2 ENSMUST00000175947.1 |
Plch1
|
phospholipase C, eta 1 |
| chr15_-_98934522 | 0.77 |
ENSMUST00000077577.7
|
Tuba1b
|
tubulin, alpha 1B |
| chr15_+_6422240 | 0.77 |
ENSMUST00000163082.1
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr10_+_116177217 | 0.75 |
ENSMUST00000148731.1
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr2_-_140671400 | 0.75 |
ENSMUST00000056760.3
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr11_-_109473220 | 0.73 |
ENSMUST00000070872.6
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr1_-_144249134 | 0.71 |
ENSMUST00000172388.1
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr8_-_47352348 | 0.70 |
ENSMUST00000110367.2
|
Stox2
|
storkhead box 2 |
| chr4_+_136284708 | 0.70 |
ENSMUST00000130223.1
|
Zfp46
|
zinc finger protein 46 |
| chr3_-_89349968 | 0.70 |
ENSMUST00000074582.6
ENSMUST00000107448.2 ENSMUST00000029676.5 |
Adam15
|
a disintegrin and metallopeptidase domain 15 (metargidin) |
| chr17_-_15826521 | 0.69 |
ENSMUST00000170578.1
|
Rgmb
|
RGM domain family, member B |
| chr1_+_164048214 | 0.69 |
ENSMUST00000027874.5
|
Sele
|
selectin, endothelial cell |
| chr11_+_98026918 | 0.69 |
ENSMUST00000017548.6
|
Rpl19
|
ribosomal protein L19 |
| chr11_+_96282529 | 0.67 |
ENSMUST00000125410.1
|
Hoxb8
|
homeobox B8 |
| chr3_-_87768966 | 0.66 |
ENSMUST00000174776.1
|
Pear1
|
platelet endothelial aggregation receptor 1 |
| chr11_+_96282648 | 0.65 |
ENSMUST00000168043.1
|
Hoxb8
|
homeobox B8 |
| chr16_+_45224315 | 0.64 |
ENSMUST00000102802.3
ENSMUST00000063654.4 |
Btla
|
B and T lymphocyte associated |
| chrX_+_150547375 | 0.64 |
ENSMUST00000066337.6
ENSMUST00000112715.1 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr3_-_19265007 | 0.64 |
ENSMUST00000091314.4
|
Pde7a
|
phosphodiesterase 7A |
| chr6_-_132957919 | 0.64 |
ENSMUST00000082085.4
|
Tas2r131
|
taste receptor, type 2, member 131 |
| chr6_-_52234902 | 0.64 |
ENSMUST00000125581.1
|
Hoxa10
|
homeobox A10 |
| chr2_-_45112890 | 0.63 |
ENSMUST00000076836.6
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_27463994 | 0.63 |
ENSMUST00000164296.1
|
Brd3
|
bromodomain containing 3 |
| chr3_-_89349939 | 0.63 |
ENSMUST00000107446.1
|
Adam15
|
a disintegrin and metallopeptidase domain 15 (metargidin) |
| chr7_-_119459266 | 0.63 |
ENSMUST00000033255.5
|
Gp2
|
glycoprotein 2 (zymogen granule membrane) |
| chr5_+_31054766 | 0.62 |
ENSMUST00000013773.5
ENSMUST00000114646.1 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chrX_-_12673540 | 0.61 |
ENSMUST00000060108.6
|
1810030O07Rik
|
RIKEN cDNA 1810030O07 gene |
| chr3_+_3634145 | 0.60 |
ENSMUST00000108394.1
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr6_-_87335758 | 0.60 |
ENSMUST00000042025.9
|
Antxr1
|
anthrax toxin receptor 1 |
| chr4_+_108479081 | 0.60 |
ENSMUST00000155068.1
|
Zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr13_-_101692624 | 0.59 |
ENSMUST00000035532.6
|
Pik3r1
|
phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha) |
| chrX_+_111168340 | 0.59 |
ENSMUST00000075226.4
|
Gm10112
|
predicted pseudogene 10112 |
| chr5_+_150259922 | 0.59 |
ENSMUST00000087204.5
|
Fry
|
furry homolog (Drosophila) |
| chr1_-_120270253 | 0.59 |
ENSMUST00000112639.1
|
Steap3
|
STEAP family member 3 |
| chr11_-_100146120 | 0.59 |
ENSMUST00000007317.7
|
Krt19
|
keratin 19 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxd11_Cdx1_Hoxc11
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 14.3 | GO:0045575 | basophil activation(GO:0045575) |
| 3.3 | 26.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 1.6 | 6.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 1.4 | 4.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 1.4 | 4.1 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 1.4 | 6.8 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 1.3 | 5.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 1.0 | 4.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 1.0 | 1.9 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.8 | 2.5 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.8 | 8.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.8 | 2.4 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.7 | 2.7 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) positive regulation of endocytic recycling(GO:2001137) |
| 0.7 | 5.3 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.7 | 10.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.6 | 1.8 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.5 | 2.7 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.4 | 3.1 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.4 | 1.3 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.4 | 1.6 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.4 | 2.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.4 | 2.6 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.4 | 1.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.4 | 1.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.3 | 1.7 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.3 | 4.8 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 0.9 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.3 | 3.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.3 | 4.0 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.3 | 9.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.3 | 3.8 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.2 | 1.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 1.2 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.2 | 3.9 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 2.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.2 | 3.0 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.2 | 2.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 1.1 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.2 | 1.4 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 0.9 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 3.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 2.0 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.2 | 1.2 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.2 | 1.2 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.2 | 2.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.2 | 2.0 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 0.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 0.5 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.2 | 5.3 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 1.8 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 2.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.0 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.6 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.1 | 1.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.6 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.5 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 | 0.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.9 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.6 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 6.1 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 5.8 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 | 0.6 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 1.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.2 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 | 1.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.3 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.1 | 1.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.9 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.4 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 1.9 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 2.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 1.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.2 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.1 | 2.9 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 8.2 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.1 | 0.6 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 2.5 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 2.2 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 5.2 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 1.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 2.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 3.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 3.5 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 2.1 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 0.6 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 1.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.5 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.9 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 4.5 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 0.2 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.1 | 0.6 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 1.1 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 2.9 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 1.1 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.0 | 1.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.5 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.7 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 1.0 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.3 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 1.9 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 1.2 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 1.3 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 1.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.8 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.1 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.0 | 0.2 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 1.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.3 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 2.7 | GO:0051928 | positive regulation of calcium ion transport(GO:0051928) |
| 0.0 | 0.1 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 1.2 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.8 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.4 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.3 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:1902572 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.1 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.6 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 1.2 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 1.3 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 1.1 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0048679 | regulation of axon regeneration(GO:0048679) |
| 0.0 | 3.2 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 1.3 | GO:0001508 | action potential(GO:0001508) |
| 0.0 | 1.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.7 | 5.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.7 | 3.9 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.6 | 2.4 | GO:0060187 | cell pole(GO:0060187) |
| 0.6 | 15.7 | GO:0032982 | myosin filament(GO:0032982) |
| 0.5 | 1.5 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.4 | 2.0 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.4 | 3.7 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.3 | 4.9 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.3 | 1.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.3 | 4.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 1.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 1.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 0.8 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.2 | 0.8 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 0.5 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.2 | 2.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 2.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 2.5 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 2.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 1.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.5 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 4.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 3.1 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 6.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.6 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 2.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.3 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 12.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 5.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.4 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 3.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.2 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 4.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.8 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 2.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 2.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 3.3 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 20.4 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 1.4 | 6.8 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.9 | 4.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.8 | 8.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.8 | 2.3 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.8 | 5.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.7 | 26.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.6 | 4.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.6 | 4.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.6 | 10.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.5 | 2.5 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.5 | 5.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.4 | 1.2 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.3 | 4.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.3 | 6.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 6.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.3 | 1.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 4.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 0.8 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.2 | 2.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 2.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 1.4 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 1.1 | GO:0034988 | mu-type opioid receptor binding(GO:0031852) Fc-gamma receptor I complex binding(GO:0034988) |
| 0.2 | 0.9 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 0.6 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.2 | 1.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 5.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.2 | 1.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 1.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 1.8 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 1.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 3.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 3.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 6.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 16.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.6 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 0.9 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.7 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 3.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.0 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 1.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.3 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.1 | 9.9 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 1.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 1.5 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 1.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.8 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 8.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 0.6 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.6 | GO:0052851 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 1.0 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.7 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 1.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.3 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 5.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 1.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 2.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 13.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 7.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.5 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.7 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 2.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 2.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 2.4 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.2 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 1.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 2.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 2.3 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.9 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 1.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 28.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.3 | 10.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 12.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 5.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 5.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 8.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 3.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 7.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 4.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 26.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 5.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.4 | 4.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 8.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 2.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 5.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 0.8 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.1 | 0.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 3.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 7.7 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 2.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 4.6 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 1.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |