Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
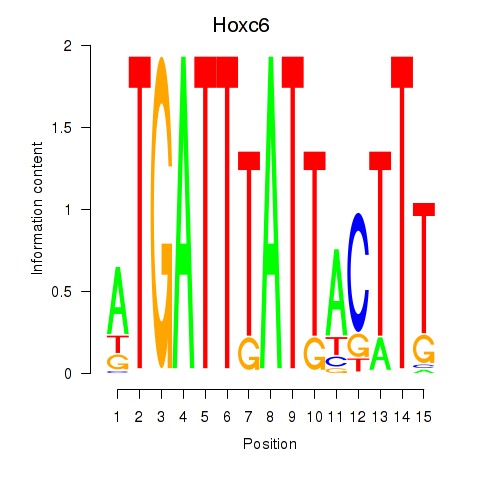
Results for Hoxc6
Z-value: 0.60
Transcription factors associated with Hoxc6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc6
|
ENSMUSG00000001661.4 | homeobox C6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxc6 | mm10_v2_chr15_+_103009479_103009507 | 0.21 | 2.1e-01 | Click! |
Activity profile of Hoxc6 motif
Sorted Z-values of Hoxc6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_113532288 | 1.60 |
ENSMUST00000132353.1
|
Amy2a1
|
amylase 2a1 |
| chr7_-_14123042 | 1.50 |
ENSMUST00000098809.2
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr3_-_145032765 | 1.49 |
ENSMUST00000029919.5
|
Clca3
|
chloride channel calcium activated 3 |
| chr14_+_27000362 | 1.09 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr3_-_92485886 | 0.78 |
ENSMUST00000054599.7
|
Sprr1a
|
small proline-rich protein 1A |
| chr11_-_109472611 | 0.65 |
ENSMUST00000168740.1
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr6_+_128399881 | 0.62 |
ENSMUST00000120405.1
|
Nrip2
|
nuclear receptor interacting protein 2 |
| chr6_-_65144908 | 0.62 |
ENSMUST00000031982.4
|
Hpgds
|
hematopoietic prostaglandin D synthase |
| chr6_+_128399766 | 0.57 |
ENSMUST00000001561.5
|
Nrip2
|
nuclear receptor interacting protein 2 |
| chr6_+_36388055 | 0.52 |
ENSMUST00000172278.1
|
Chrm2
|
cholinergic receptor, muscarinic 2, cardiac |
| chr3_+_146150174 | 0.51 |
ENSMUST00000098524.4
|
Mcoln2
|
mucolipin 2 |
| chr10_-_61784014 | 0.49 |
ENSMUST00000020283.4
|
H2afy2
|
H2A histone family, member Y2 |
| chrX_-_94212638 | 0.48 |
ENSMUST00000113922.1
|
Eif2s3x
|
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
| chr5_+_24413406 | 0.47 |
ENSMUST00000049346.5
|
Asic3
|
acid-sensing (proton-gated) ion channel 3 |
| chrX_+_49463926 | 0.45 |
ENSMUST00000130558.1
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr13_+_93304066 | 0.44 |
ENSMUST00000109493.1
|
Homer1
|
homer homolog 1 (Drosophila) |
| chr13_+_93304799 | 0.43 |
ENSMUST00000080127.5
|
Homer1
|
homer homolog 1 (Drosophila) |
| chr15_+_91673175 | 0.42 |
ENSMUST00000060642.6
|
Lrrk2
|
leucine-rich repeat kinase 2 |
| chr16_-_21995478 | 0.41 |
ENSMUST00000074230.4
ENSMUST00000060673.6 |
Liph
|
lipase, member H |
| chr10_-_100589205 | 0.39 |
ENSMUST00000054471.8
|
4930430F08Rik
|
RIKEN cDNA 4930430F08 gene |
| chr7_+_45554893 | 0.37 |
ENSMUST00000107752.3
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr1_-_138175238 | 0.37 |
ENSMUST00000182536.1
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr7_-_115846080 | 0.36 |
ENSMUST00000166207.1
|
Sox6
|
SRY-box containing gene 6 |
| chr6_+_52177498 | 0.35 |
ENSMUST00000070587.3
|
5730596B20Rik
|
RIKEN cDNA 5730596B20 gene |
| chr1_-_138175283 | 0.35 |
ENSMUST00000182755.1
ENSMUST00000183262.1 ENSMUST00000027645.7 ENSMUST00000112036.2 ENSMUST00000182283.1 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr1_-_138175126 | 0.33 |
ENSMUST00000183301.1
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr19_-_11660516 | 0.32 |
ENSMUST00000135994.1
ENSMUST00000121793.1 ENSMUST00000069681.3 |
Plac1l
|
placenta-specific 1-like |
| chrX_-_94212685 | 0.31 |
ENSMUST00000050328.8
|
Eif2s3x
|
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
| chr11_-_94782500 | 0.30 |
ENSMUST00000162809.2
|
Tmem92
|
transmembrane protein 92 |
| chr11_-_94782703 | 0.30 |
ENSMUST00000100554.1
|
Tmem92
|
transmembrane protein 92 |
| chr19_+_11469353 | 0.28 |
ENSMUST00000165310.1
|
Ms4a6c
|
membrane-spanning 4-domains, subfamily A, member 6C |
| chr18_+_53551594 | 0.27 |
ENSMUST00000115398.1
|
Prdm6
|
PR domain containing 6 |
| chr6_-_141773810 | 0.27 |
ENSMUST00000148411.1
|
Gm5724
|
predicted gene 5724 |
| chr2_+_150786735 | 0.23 |
ENSMUST00000045441.7
|
Pygb
|
brain glycogen phosphorylase |
| chr15_+_44619551 | 0.23 |
ENSMUST00000022964.7
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chrX_+_164162167 | 0.23 |
ENSMUST00000131543.1
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr16_-_58718724 | 0.22 |
ENSMUST00000089318.3
|
Gpr15
|
G protein-coupled receptor 15 |
| chr5_-_110046486 | 0.22 |
ENSMUST00000167969.1
|
Gm17655
|
predicted gene, 17655 |
| chr7_-_24299310 | 0.21 |
ENSMUST00000145131.1
|
Zfp61
|
zinc finger protein 61 |
| chr3_+_5218516 | 0.21 |
ENSMUST00000175866.1
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr1_-_22315792 | 0.21 |
ENSMUST00000164877.1
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr4_+_132351768 | 0.21 |
ENSMUST00000172202.1
|
Gm17300
|
predicted gene, 17300 |
| chr9_+_107551516 | 0.21 |
ENSMUST00000093786.2
ENSMUST00000122225.1 |
Rassf1
|
Ras association (RalGDS/AF-6) domain family member 1 |
| chr4_-_132351636 | 0.21 |
ENSMUST00000105951.1
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr3_+_136670076 | 0.21 |
ENSMUST00000070198.7
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform |
| chr4_-_147642496 | 0.20 |
ENSMUST00000133006.1
ENSMUST00000037565.7 ENSMUST00000105720.1 |
2610305D13Rik
|
RIKEN cDNA 2610305D13 gene |
| chr15_+_100154379 | 0.20 |
ENSMUST00000023768.6
ENSMUST00000108971.2 |
Dip2b
|
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
| chr3_+_151437887 | 0.20 |
ENSMUST00000046977.7
|
Eltd1
|
EGF, latrophilin seven transmembrane domain containing 1 |
| chr10_+_37139558 | 0.20 |
ENSMUST00000062667.3
|
5930403N24Rik
|
RIKEN cDNA 5930403N24 gene |
| chr5_+_17574268 | 0.19 |
ENSMUST00000030568.7
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr15_-_50889043 | 0.18 |
ENSMUST00000183997.1
ENSMUST00000183757.1 |
Trps1
|
trichorhinophalangeal syndrome I (human) |
| chr3_+_66985680 | 0.18 |
ENSMUST00000065047.6
|
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr3_+_66985647 | 0.16 |
ENSMUST00000162362.1
ENSMUST00000065074.7 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr3_+_66985700 | 0.16 |
ENSMUST00000046542.6
ENSMUST00000162693.1 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr10_-_70655934 | 0.16 |
ENSMUST00000162144.1
ENSMUST00000162793.1 |
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like |
| chr7_+_45554937 | 0.16 |
ENSMUST00000133242.3
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr3_+_129213920 | 0.15 |
ENSMUST00000042587.10
|
Pitx2
|
paired-like homeodomain transcription factor 2 |
| chr11_-_23895208 | 0.15 |
ENSMUST00000102863.2
ENSMUST00000020513.3 |
Papolg
|
poly(A) polymerase gamma |
| chr11_-_111800870 | 0.14 |
ENSMUST00000124855.1
|
Gm11674
|
predicted gene 11674 |
| chr15_-_50889691 | 0.14 |
ENSMUST00000165201.2
ENSMUST00000184458.1 |
Trps1
|
trichorhinophalangeal syndrome I (human) |
| chr14_+_53845234 | 0.13 |
ENSMUST00000103674.4
|
Trav19
|
T cell receptor alpha variable 19 |
| chr7_-_16285454 | 0.13 |
ENSMUST00000174270.1
|
Ccdc9
|
coiled-coil domain containing 9 |
| chr4_+_107707030 | 0.12 |
ENSMUST00000140321.1
|
1700047F07Rik
|
RIKEN cDNA 1700047F07 gene |
| chr6_-_53978662 | 0.12 |
ENSMUST00000166545.1
|
Cpvl
|
carboxypeptidase, vitellogenic-like |
| chr9_-_27155418 | 0.12 |
ENSMUST00000167074.1
ENSMUST00000034472.8 |
Jam3
|
junction adhesion molecule 3 |
| chr1_+_19208914 | 0.12 |
ENSMUST00000027059.4
|
Tfap2b
|
transcription factor AP-2 beta |
| chr17_-_35115428 | 0.12 |
ENSMUST00000172854.1
ENSMUST00000062657.4 |
Ly6g5b
|
lymphocyte antigen 6 complex, locus G5B |
| chr3_+_5218546 | 0.11 |
ENSMUST00000026284.6
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr6_-_3494587 | 0.10 |
ENSMUST00000049985.8
|
Hepacam2
|
HEPACAM family member 2 |
| chr10_+_112083345 | 0.10 |
ENSMUST00000148897.1
ENSMUST00000020434.3 |
Glipr1l2
|
GLI pathogenesis-related 1 like 2 |
| chr8_-_9976294 | 0.10 |
ENSMUST00000095476.4
|
Lig4
|
ligase IV, DNA, ATP-dependent |
| chr7_+_123123870 | 0.09 |
ENSMUST00000094053.5
|
Tnrc6a
|
trinucleotide repeat containing 6a |
| chr4_+_107707056 | 0.09 |
ENSMUST00000147654.1
|
1700047F07Rik
|
RIKEN cDNA 1700047F07 gene |
| chr7_+_83148644 | 0.09 |
ENSMUST00000066005.5
|
A530021J07Rik
|
Riken cDNA A530021J07 gene |
| chr2_-_51149100 | 0.09 |
ENSMUST00000154545.1
ENSMUST00000017288.2 |
Rnd3
|
Rho family GTPase 3 |
| chr17_+_12876034 | 0.08 |
ENSMUST00000075296.2
|
Mrgprh
|
MAS-related GPR, member H |
| chr10_-_107719978 | 0.08 |
ENSMUST00000050702.7
|
Ptprq
|
protein tyrosine phosphatase, receptor type, Q |
| chr18_+_56572822 | 0.08 |
ENSMUST00000008445.5
|
Phax
|
phosphorylated adaptor for RNA export |
| chr19_+_6364557 | 0.06 |
ENSMUST00000155973.1
|
Sf1
|
splicing factor 1 |
| chr6_-_129917650 | 0.06 |
ENSMUST00000118060.1
|
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chr3_+_5218589 | 0.06 |
ENSMUST00000177488.1
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr7_-_19604444 | 0.05 |
ENSMUST00000086041.5
|
Clasrp
|
CLK4-associating serine/arginine rich protein |
| chr4_+_99656299 | 0.04 |
ENSMUST00000087285.3
|
Foxd3
|
forkhead box D3 |
| chr5_-_23616528 | 0.03 |
ENSMUST00000088392.4
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr19_+_11518493 | 0.03 |
ENSMUST00000025580.3
|
Ms4a6b
|
membrane-spanning 4-domains, subfamily A, member 6B |
| chr7_+_5020376 | 0.02 |
ENSMUST00000076251.4
|
Zfp865
|
zinc finger protein 865 |
| chrX_+_102802963 | 0.02 |
ENSMUST00000118218.1
|
Dmrtc1c1
|
DMRT-like family C1c1 |
| chr9_-_21760275 | 0.01 |
ENSMUST00000098942.4
|
Spc24
|
SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr13_-_64903888 | 0.01 |
ENSMUST00000091554.4
|
Cntnap3
|
contactin associated protein-like 3 |
| chr11_-_24075054 | 0.01 |
ENSMUST00000068360.1
|
A830031A19Rik
|
RIKEN cDNA A830031A19 gene |
| chr4_+_33062999 | 0.00 |
ENSMUST00000108162.1
ENSMUST00000024035.2 |
Gabrr2
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 2 |
| chr16_-_56712825 | 0.00 |
ENSMUST00000136394.1
|
Tfg
|
Trk-fused gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxc6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:2000473 | immunoglobulin biosynthetic process(GO:0002378) regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.1 | 0.4 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.6 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 1.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 0.5 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.5 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.5 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 | 0.2 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 0.9 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.2 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.1 | 0.2 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 0.3 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.1 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.0 | 0.4 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.8 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.5 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.3 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.1 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.5 | GO:0046677 | response to antibiotic(GO:0046677) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 0.8 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.5 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.2 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.9 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.2 | 0.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 0.6 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.4 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.1 | 0.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.5 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 0.9 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 0.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 1.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 1.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |