Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Hoxa10
Z-value: 1.51
Transcription factors associated with Hoxa10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa10
|
ENSMUSG00000000938.11 | homeobox A10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa10 | mm10_v2_chr6_-_52234902_52234960 | -0.39 | 1.9e-02 | Click! |
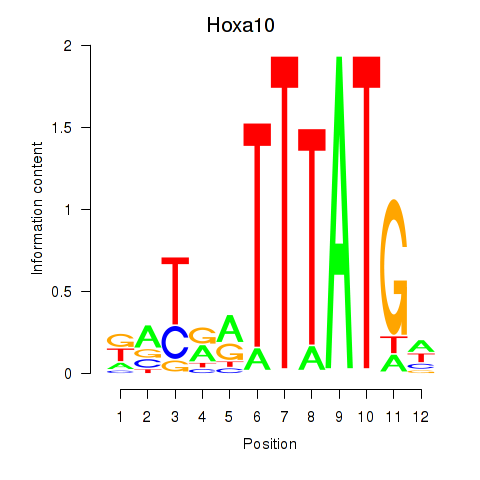
Activity profile of Hoxa10 motif
Sorted Z-values of Hoxa10 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_102658640 | 6.07 |
ENSMUST00000080210.3
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr2_+_68117713 | 4.76 |
ENSMUST00000112346.2
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr7_-_14446651 | 4.71 |
ENSMUST00000125941.1
|
2810007J24Rik
|
RIKEN cDNA 2810007J24 gene |
| chr5_-_87092546 | 4.16 |
ENSMUST00000132667.1
ENSMUST00000145617.1 ENSMUST00000094649.4 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr5_+_146079254 | 4.15 |
ENSMUST00000035571.8
|
Cyp3a59
|
cytochrome P450, family 3, subfamily a, polypeptide 59 |
| chr13_-_22219820 | 4.05 |
ENSMUST00000057516.1
|
Vmn1r193
|
vomeronasal 1 receptor 193 |
| chr5_-_87091150 | 3.94 |
ENSMUST00000154455.1
|
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr13_-_41847482 | 3.65 |
ENSMUST00000072012.3
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr13_-_41847599 | 3.40 |
ENSMUST00000179758.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr2_-_164638789 | 2.99 |
ENSMUST00000109336.1
|
Wfdc16
|
WAP four-disulfide core domain 16 |
| chr1_-_140183283 | 2.97 |
ENSMUST00000111977.1
|
Cfh
|
complement component factor h |
| chr9_+_55326913 | 2.96 |
ENSMUST00000085754.3
ENSMUST00000034862.4 |
AI118078
|
expressed sequence AI118078 |
| chr1_+_167618246 | 2.87 |
ENSMUST00000111380.1
|
Rxrg
|
retinoid X receptor gamma |
| chr1_+_74713551 | 2.64 |
ENSMUST00000027356.5
|
Cyp27a1
|
cytochrome P450, family 27, subfamily a, polypeptide 1 |
| chr19_+_4711153 | 2.62 |
ENSMUST00000008991.6
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr7_+_51880312 | 2.61 |
ENSMUST00000145049.1
|
Gas2
|
growth arrest specific 2 |
| chr19_+_30232921 | 2.58 |
ENSMUST00000025797.5
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chr1_-_140183404 | 2.50 |
ENSMUST00000066859.6
ENSMUST00000111976.2 |
Cfh
|
complement component factor h |
| chr16_+_56477838 | 2.40 |
ENSMUST00000048471.7
ENSMUST00000096013.3 ENSMUST00000096012.3 ENSMUST00000171000.1 |
Abi3bp
|
ABI gene family, member 3 (NESH) binding protein |
| chr17_+_36958623 | 2.31 |
ENSMUST00000173814.1
|
Znrd1as
|
Znrd1 antisense |
| chr17_+_36958571 | 2.31 |
ENSMUST00000040177.6
|
Znrd1as
|
Znrd1 antisense |
| chr15_+_4727265 | 2.19 |
ENSMUST00000162350.1
|
C6
|
complement component 6 |
| chr4_+_42629719 | 2.17 |
ENSMUST00000166898.2
|
Gm2564
|
predicted gene 2564 |
| chr4_-_119383732 | 2.11 |
ENSMUST00000044781.2
ENSMUST00000084307.3 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr6_-_83677807 | 2.10 |
ENSMUST00000037882.6
|
Cd207
|
CD207 antigen |
| chr3_+_129836729 | 2.00 |
ENSMUST00000077918.5
|
Cfi
|
complement component factor i |
| chr14_-_122913751 | 1.96 |
ENSMUST00000160401.1
|
Ggact
|
gamma-glutamylamine cyclotransferase |
| chr6_+_59208870 | 1.90 |
ENSMUST00000062626.3
|
Tigd2
|
tigger transposable element derived 2 |
| chrX_+_10252361 | 1.89 |
ENSMUST00000115528.2
|
Otc
|
ornithine transcarbamylase |
| chr2_+_91255954 | 1.88 |
ENSMUST00000134699.1
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr18_-_12860197 | 1.86 |
ENSMUST00000124570.1
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chrX_+_10252305 | 1.81 |
ENSMUST00000049910.6
|
Otc
|
ornithine transcarbamylase |
| chr9_-_51328898 | 1.79 |
ENSMUST00000039959.4
|
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr1_+_127729405 | 1.78 |
ENSMUST00000038006.6
|
Acmsd
|
amino carboxymuconate semialdehyde decarboxylase |
| chr4_-_109202217 | 1.75 |
ENSMUST00000160774.1
ENSMUST00000030288.7 ENSMUST00000162787.2 |
Osbpl9
|
oxysterol binding protein-like 9 |
| chr7_-_73537621 | 1.74 |
ENSMUST00000172704.1
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr18_-_3309723 | 1.70 |
ENSMUST00000136961.1
ENSMUST00000152108.1 |
Crem
|
cAMP responsive element modulator |
| chr18_-_3281712 | 1.68 |
ENSMUST00000182204.1
ENSMUST00000154705.1 ENSMUST00000182833.1 ENSMUST00000151084.1 |
Crem
|
cAMP responsive element modulator |
| chr13_+_23839401 | 1.68 |
ENSMUST00000039721.7
ENSMUST00000166467.1 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr2_+_91256144 | 1.67 |
ENSMUST00000154959.1
ENSMUST00000059566.4 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr15_-_3303521 | 1.63 |
ENSMUST00000165386.1
|
Ccdc152
|
coiled-coil domain containing 152 |
| chr13_+_23839445 | 1.59 |
ENSMUST00000091698.4
ENSMUST00000110422.1 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr6_+_54264839 | 1.58 |
ENSMUST00000146114.1
|
Chn2
|
chimerin (chimaerin) 2 |
| chr2_+_83812567 | 1.55 |
ENSMUST00000051454.3
|
Fam171b
|
family with sequence similarity 171, member B |
| chr5_-_123666682 | 1.53 |
ENSMUST00000149410.1
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr7_+_45277502 | 1.53 |
ENSMUST00000176276.1
|
1700039E15Rik
|
RIKEN cDNA 1700039E15 gene |
| chr15_+_3270767 | 1.43 |
ENSMUST00000082424.4
ENSMUST00000159158.1 ENSMUST00000159216.1 ENSMUST00000160311.1 |
Sepp1
|
selenoprotein P, plasma, 1 |
| chr3_+_94398517 | 1.41 |
ENSMUST00000050975.3
|
Lingo4
|
leucine rich repeat and Ig domain containing 4 |
| chr18_-_39487096 | 1.38 |
ENSMUST00000097592.2
ENSMUST00000115571.1 |
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr13_-_90089060 | 1.37 |
ENSMUST00000161396.1
|
Xrcc4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr9_+_54538984 | 1.33 |
ENSMUST00000060242.5
ENSMUST00000118413.1 |
Sh2d7
|
SH2 domain containing 7 |
| chr19_+_31868754 | 1.31 |
ENSMUST00000075838.5
|
A1cf
|
APOBEC1 complementation factor |
| chr19_+_29925107 | 1.30 |
ENSMUST00000120388.2
ENSMUST00000144528.1 ENSMUST00000177518.1 |
Il33
|
interleukin 33 |
| chr2_+_15049395 | 1.29 |
ENSMUST00000017562.6
|
Arl5b
|
ADP-ribosylation factor-like 5B |
| chr3_+_132634725 | 1.29 |
ENSMUST00000163241.1
|
Gm5549
|
predicted gene 5549 |
| chr18_+_33464163 | 1.27 |
ENSMUST00000097634.3
|
Gm10549
|
predicted gene 10549 |
| chrM_+_14138 | 1.27 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr18_-_3281752 | 1.26 |
ENSMUST00000140332.1
ENSMUST00000147138.1 |
Crem
|
cAMP responsive element modulator |
| chrM_+_3906 | 1.26 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chrM_-_14060 | 1.23 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr15_-_54919961 | 1.21 |
ENSMUST00000167541.2
ENSMUST00000041591.9 ENSMUST00000173516.1 |
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr18_-_3309858 | 1.19 |
ENSMUST00000144496.1
ENSMUST00000154715.1 |
Crem
|
cAMP responsive element modulator |
| chr2_-_77519565 | 1.18 |
ENSMUST00000111830.2
|
Zfp385b
|
zinc finger protein 385B |
| chr17_-_6079693 | 1.17 |
ENSMUST00000024570.5
ENSMUST00000097432.3 |
Serac1
|
serine active site containing 1 |
| chr4_-_138326234 | 1.16 |
ENSMUST00000105817.3
ENSMUST00000030536.6 |
Pink1
|
PTEN induced putative kinase 1 |
| chr9_-_50739365 | 1.14 |
ENSMUST00000117093.1
ENSMUST00000121634.1 |
Dixdc1
|
DIX domain containing 1 |
| chr15_-_54920115 | 1.13 |
ENSMUST00000171545.1
|
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr2_-_51149100 | 1.12 |
ENSMUST00000154545.1
ENSMUST00000017288.2 |
Rnd3
|
Rho family GTPase 3 |
| chr9_+_122888471 | 1.11 |
ENSMUST00000063980.6
|
Zkscan7
|
zinc finger with KRAB and SCAN domains 7 |
| chr6_-_142278836 | 1.07 |
ENSMUST00000111825.3
|
Slco1a5
|
solute carrier organic anion transporter family, member 1a5 |
| chr10_-_56228636 | 1.07 |
ENSMUST00000099739.3
|
Tbc1d32
|
TBC1 domain family, member 32 |
| chr2_+_177508570 | 1.07 |
ENSMUST00000108940.2
|
Gm14403
|
predicted gene 14403 |
| chr13_-_74376566 | 1.06 |
ENSMUST00000091481.2
|
Zfp72
|
zinc finger protein 72 |
| chr17_+_88626549 | 1.03 |
ENSMUST00000163588.1
ENSMUST00000064035.6 |
Ston1
|
stonin 1 |
| chr8_-_41054771 | 0.98 |
ENSMUST00000093534.4
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr16_-_35871544 | 0.97 |
ENSMUST00000042665.8
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr4_+_141746672 | 0.95 |
ENSMUST00000038161.4
|
Agmat
|
agmatine ureohydrolase (agmatinase) |
| chr17_+_88626569 | 0.92 |
ENSMUST00000150023.1
|
Ston1
|
stonin 1 |
| chr2_+_69790968 | 0.89 |
ENSMUST00000180290.1
|
Phospho2
|
phosphatase, orphan 2 |
| chr3_+_40540751 | 0.89 |
ENSMUST00000091186.3
|
Intu
|
inturned planar cell polarity effector homolog (Drosophila) |
| chr15_-_58847321 | 0.86 |
ENSMUST00000075109.5
|
Ube2d4
|
ubiquitin-conjugating enzyme E2D 4 |
| chr1_+_174050375 | 0.85 |
ENSMUST00000062665.3
|
Olfr432
|
olfactory receptor 432 |
| chr14_+_41131777 | 0.83 |
ENSMUST00000022314.3
ENSMUST00000170719.1 |
Sftpa1
|
surfactant associated protein A1 |
| chr1_+_193173469 | 0.81 |
ENSMUST00000161235.1
ENSMUST00000110831.2 ENSMUST00000178744.1 |
A130010J15Rik
|
RIKEN cDNA A130010J15 gene |
| chr1_+_182124737 | 0.81 |
ENSMUST00000111018.1
ENSMUST00000027792.5 |
Srp9
|
signal recognition particle 9 |
| chr8_+_106935720 | 0.80 |
ENSMUST00000047425.3
|
Sntb2
|
syntrophin, basic 2 |
| chr12_+_10390756 | 0.76 |
ENSMUST00000020947.5
|
Rdh14
|
retinol dehydrogenase 14 (all-trans and 9-cis) |
| chr2_+_101624696 | 0.75 |
ENSMUST00000044031.3
|
Rag2
|
recombination activating gene 2 |
| chr11_-_100762928 | 0.74 |
ENSMUST00000107360.2
ENSMUST00000055083.3 |
Hcrt
|
hypocretin |
| chr3_-_59262825 | 0.73 |
ENSMUST00000050360.7
|
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr8_+_46492789 | 0.72 |
ENSMUST00000110371.1
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr17_+_13061099 | 0.72 |
ENSMUST00000086787.4
ENSMUST00000116666.1 |
Tcp10b
|
t-complex protein 10b |
| chr2_-_91255995 | 0.72 |
ENSMUST00000180732.1
|
Gm17281
|
predicted gene, 17281 |
| chr3_-_75270073 | 0.71 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr9_+_13765970 | 0.70 |
ENSMUST00000152532.1
|
Mtmr2
|
myotubularin related protein 2 |
| chr7_+_29238434 | 0.70 |
ENSMUST00000108237.1
|
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr6_-_122856151 | 0.70 |
ENSMUST00000042081.8
|
C3ar1
|
complement component 3a receptor 1 |
| chr17_+_36121666 | 0.69 |
ENSMUST00000173128.1
|
Gm19684
|
predicted gene, 19684 |
| chr10_+_29143996 | 0.67 |
ENSMUST00000092629.2
|
Soga3
|
SOGA family member 3 |
| chr11_+_87582201 | 0.66 |
ENSMUST00000133202.1
|
Sept4
|
septin 4 |
| chr8_+_46986913 | 0.65 |
ENSMUST00000039840.7
ENSMUST00000119686.1 |
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chrX_+_139086226 | 0.64 |
ENSMUST00000033625.1
|
4930513O06Rik
|
RIKEN cDNA 4930513O06 gene |
| chr6_+_6955993 | 0.64 |
ENSMUST00000040826.5
|
Acn9
|
ACN9 homolog (S. cerevisiae) |
| chr9_+_62342449 | 0.64 |
ENSMUST00000156461.1
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chrX_-_136741155 | 0.64 |
ENSMUST00000166930.1
ENSMUST00000113095.1 ENSMUST00000155207.1 ENSMUST00000080411.6 ENSMUST00000169418.1 |
Morf4l2
|
mortality factor 4 like 2 |
| chr19_+_24875679 | 0.63 |
ENSMUST00000073080.5
|
Gm10053
|
predicted gene 10053 |
| chr3_-_94436574 | 0.62 |
ENSMUST00000029787.4
|
Oaz3
|
ornithine decarboxylase antizyme 3 |
| chr5_+_145345254 | 0.59 |
ENSMUST00000079268.7
|
Cyp3a57
|
cytochrome P450, family 3, subfamily a, polypeptide 57 |
| chr1_+_153874335 | 0.58 |
ENSMUST00000055314.3
|
Gm5531
|
predicted gene 5531 |
| chr1_+_179961110 | 0.58 |
ENSMUST00000076687.5
ENSMUST00000097450.3 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr3_+_106486009 | 0.56 |
ENSMUST00000183271.1
ENSMUST00000061206.3 |
Dennd2d
|
DENN/MADD domain containing 2D |
| chr7_+_102229999 | 0.56 |
ENSMUST00000120119.1
|
Pgap2
|
post-GPI attachment to proteins 2 |
| chr6_-_24528013 | 0.54 |
ENSMUST00000023851.5
|
Ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chrX_+_103356464 | 0.54 |
ENSMUST00000116547.2
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr7_+_29238323 | 0.53 |
ENSMUST00000108238.1
ENSMUST00000032809.3 ENSMUST00000138128.1 |
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr13_-_92030897 | 0.53 |
ENSMUST00000149630.1
|
Rasgrf2
|
RAS protein-specific guanine nucleotide-releasing factor 2 |
| chr5_+_121802230 | 0.53 |
ENSMUST00000162995.1
|
Atxn2
|
ataxin 2 |
| chr17_+_33824591 | 0.51 |
ENSMUST00000048249.6
|
Ndufa7
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7 (B14.5a) |
| chr1_-_136131171 | 0.51 |
ENSMUST00000146091.3
ENSMUST00000165464.1 ENSMUST00000166747.1 ENSMUST00000134998.1 |
Gm15850
|
predicted gene 15850 |
| chr5_+_121749196 | 0.50 |
ENSMUST00000161064.1
|
Atxn2
|
ataxin 2 |
| chr10_-_11082287 | 0.50 |
ENSMUST00000105561.2
ENSMUST00000044306.6 |
Grm1
|
glutamate receptor, metabotropic 1 |
| chr2_-_114201416 | 0.50 |
ENSMUST00000050668.3
|
Zfp770
|
zinc finger protein 770 |
| chr18_+_37513652 | 0.49 |
ENSMUST00000061405.4
|
Pcdhb21
|
protocadherin beta 21 |
| chr6_+_21949571 | 0.49 |
ENSMUST00000031680.3
ENSMUST00000115389.1 ENSMUST00000151473.1 |
Ing3
|
inhibitor of growth family, member 3 |
| chr6_-_129678908 | 0.47 |
ENSMUST00000118447.1
ENSMUST00000169545.1 ENSMUST00000032270.6 ENSMUST00000032271.6 |
Klrc1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr11_-_78984946 | 0.47 |
ENSMUST00000108268.3
|
Lgals9
|
lectin, galactose binding, soluble 9 |
| chr19_+_11965817 | 0.47 |
ENSMUST00000025590.9
|
Osbp
|
oxysterol binding protein |
| chr2_-_79908428 | 0.44 |
ENSMUST00000102652.3
ENSMUST00000102651.3 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr11_-_78984831 | 0.44 |
ENSMUST00000073001.4
ENSMUST00000108269.3 |
Lgals9
|
lectin, galactose binding, soluble 9 |
| chr14_+_52821860 | 0.43 |
ENSMUST00000181793.1
ENSMUST00000184436.1 |
Trav6d-6
|
T cell receptor alpha variable 6D-6 |
| chr12_+_55239326 | 0.43 |
ENSMUST00000164243.1
|
Srp54c
|
signal recognition particle 54C |
| chr9_+_39993812 | 0.42 |
ENSMUST00000057161.3
|
Olfr978
|
olfactory receptor 978 |
| chr5_+_121777929 | 0.41 |
ENSMUST00000160821.1
|
Atxn2
|
ataxin 2 |
| chr8_+_85432686 | 0.41 |
ENSMUST00000180883.1
|
1700051O22Rik
|
RIKEN cDNA 1700051O22 Gene |
| chr9_+_38719024 | 0.41 |
ENSMUST00000129598.1
|
Vwa5a
|
von Willebrand factor A domain containing 5A |
| chr10_-_29362012 | 0.40 |
ENSMUST00000161508.1
|
Rnf146
|
ring finger protein 146 |
| chr11_-_107348130 | 0.40 |
ENSMUST00000134763.1
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr9_+_58582397 | 0.39 |
ENSMUST00000176557.1
ENSMUST00000114121.4 ENSMUST00000177064.1 |
Nptn
|
neuroplastin |
| chr4_-_14796052 | 0.38 |
ENSMUST00000108276.1
ENSMUST00000023917.1 |
Lrrc69
|
leucine rich repeat containing 69 |
| chr13_+_18717289 | 0.38 |
ENSMUST00000072961.4
|
Vps41
|
vacuolar protein sorting 41 (yeast) |
| chr13_+_65512678 | 0.38 |
ENSMUST00000081471.2
|
Gm10139
|
predicted gene 10139 |
| chr3_+_93826020 | 0.37 |
ENSMUST00000081780.3
|
Tdpoz3
|
TD and POZ domain containing 3 |
| chr12_+_55089202 | 0.37 |
ENSMUST00000021407.10
|
Srp54a
|
signal recognition particle 54A |
| chr1_-_59003443 | 0.37 |
ENSMUST00000054653.6
|
Als2cr11
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 11 (human) |
| chr11_-_102579071 | 0.37 |
ENSMUST00000107080.1
|
Gm11627
|
predicted gene 11627 |
| chr12_+_52516077 | 0.37 |
ENSMUST00000110725.1
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chrX_+_99042581 | 0.36 |
ENSMUST00000036606.7
|
Stard8
|
START domain containing 8 |
| chr1_+_165769392 | 0.35 |
ENSMUST00000040298.4
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr2_-_152464612 | 0.35 |
ENSMUST00000099206.2
|
Defb23
|
defensin beta 23 |
| chr11_-_110168073 | 0.35 |
ENSMUST00000044850.3
|
Abca9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr16_-_10788649 | 0.35 |
ENSMUST00000051297.7
|
Tnp2
|
transition protein 2 |
| chr18_+_37411674 | 0.33 |
ENSMUST00000051126.2
|
Pcdhb10
|
protocadherin beta 10 |
| chr11_-_99422252 | 0.33 |
ENSMUST00000017741.3
|
Krt12
|
keratin 12 |
| chr14_-_51146757 | 0.33 |
ENSMUST00000080126.2
|
Rnase1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr3_-_30509681 | 0.32 |
ENSMUST00000173495.1
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr4_+_43406435 | 0.31 |
ENSMUST00000098106.2
ENSMUST00000139198.1 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr3_+_85915722 | 0.31 |
ENSMUST00000054148.7
|
Gm9790
|
predicted gene 9790 |
| chrX_+_56609843 | 0.30 |
ENSMUST00000077741.5
ENSMUST00000114784.2 |
Slc9a6
|
solute carrier family 9 (sodium/hydrogen exchanger), member 6 |
| chr2_-_37647199 | 0.30 |
ENSMUST00000028279.3
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr11_+_114091404 | 0.30 |
ENSMUST00000129365.1
|
1700025D23Rik
|
RIKEN cDNA 1700025D23 gene |
| chr18_-_23038656 | 0.29 |
ENSMUST00000081423.6
|
Nol4
|
nucleolar protein 4 |
| chr5_+_64160207 | 0.28 |
ENSMUST00000101195.2
|
Tbc1d1
|
TBC1 domain family, member 1 |
| chr3_-_57301919 | 0.28 |
ENSMUST00000029376.8
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr13_+_23695791 | 0.28 |
ENSMUST00000041052.2
|
Hist1h1t
|
histone cluster 1, H1t |
| chr3_+_66985680 | 0.27 |
ENSMUST00000065047.6
|
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr4_-_91376433 | 0.26 |
ENSMUST00000107109.2
ENSMUST00000107111.2 ENSMUST00000107120.1 |
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) |
| chr2_-_89752826 | 0.26 |
ENSMUST00000099765.1
|
Olfr1253
|
olfactory receptor 1253 |
| chr2_-_79908389 | 0.26 |
ENSMUST00000090756.4
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr3_-_30509752 | 0.25 |
ENSMUST00000172697.1
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr8_+_117701937 | 0.25 |
ENSMUST00000034304.7
|
Hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr1_+_179960472 | 0.25 |
ENSMUST00000097453.2
ENSMUST00000111117.1 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr17_-_25844417 | 0.25 |
ENSMUST00000176591.1
|
Rhot2
|
ras homolog gene family, member T2 |
| chr2_-_172454742 | 0.25 |
ENSMUST00000161334.1
|
Gcnt7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr11_+_71749914 | 0.24 |
ENSMUST00000150531.1
|
Wscd1
|
WSC domain containing 1 |
| chr2_-_77946375 | 0.23 |
ENSMUST00000065889.3
|
Cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr3_+_67892189 | 0.23 |
ENSMUST00000063263.3
|
Iqcj
|
IQ motif containing J |
| chr2_-_77946331 | 0.22 |
ENSMUST00000111821.2
ENSMUST00000111818.1 |
Cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr1_+_164503306 | 0.21 |
ENSMUST00000181831.1
|
Gm26685
|
predicted gene, 26685 |
| chr12_-_72917872 | 0.20 |
ENSMUST00000044000.5
|
4930447C04Rik
|
RIKEN cDNA 4930447C04 gene |
| chr5_-_21701332 | 0.19 |
ENSMUST00000115217.1
ENSMUST00000060899.8 |
Napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr1_-_156718894 | 0.18 |
ENSMUST00000086153.6
|
Fam20b
|
family with sequence similarity 20, member B |
| chr17_+_6079786 | 0.17 |
ENSMUST00000039487.3
|
Gtf2h5
|
general transcription factor IIH, polypeptide 5 |
| chr5_+_24394388 | 0.17 |
ENSMUST00000115074.1
|
Abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr1_-_132229767 | 0.15 |
ENSMUST00000086544.4
|
Gm10188
|
predicted gene 10188 |
| chr11_-_69795930 | 0.15 |
ENSMUST00000045971.8
|
Chrnb1
|
cholinergic receptor, nicotinic, beta polypeptide 1 (muscle) |
| chr19_+_39060998 | 0.15 |
ENSMUST00000087236.4
|
Cyp2c65
|
cytochrome P450, family 2, subfamily c, polypeptide 65 |
| chr9_-_100506844 | 0.13 |
ENSMUST00000112874.3
|
Nck1
|
non-catalytic region of tyrosine kinase adaptor protein 1 |
| chr6_+_65590382 | 0.12 |
ENSMUST00000114236.1
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr13_-_18031616 | 0.11 |
ENSMUST00000099736.2
|
Vdac3-ps1
|
voltage-dependent anion channel 3, pseudogene 1 |
| chr2_+_31759993 | 0.11 |
ENSMUST00000124089.1
|
Abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr8_+_110618577 | 0.10 |
ENSMUST00000034190.9
|
Vac14
|
Vac14 homolog (S. cerevisiae) |
| chr10_+_24076500 | 0.10 |
ENSMUST00000051133.5
|
Taar8a
|
trace amine-associated receptor 8A |
| chr3_+_102010138 | 0.10 |
ENSMUST00000066187.4
|
Nhlh2
|
nescient helix loop helix 2 |
| chr6_-_67491849 | 0.09 |
ENSMUST00000118364.1
|
Il23r
|
interleukin 23 receptor |
| chr9_-_39957069 | 0.08 |
ENSMUST00000169307.2
|
Olfr976
|
olfactory receptor 976 |
| chr2_-_34826187 | 0.08 |
ENSMUST00000113075.1
ENSMUST00000113080.2 ENSMUST00000091020.3 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr19_-_45591820 | 0.08 |
ENSMUST00000160003.1
ENSMUST00000162879.1 |
Fbxw4
|
F-box and WD-40 domain protein 4 |
| chr3_+_94643112 | 0.08 |
ENSMUST00000107276.1
|
Gm10972
|
predicted gene 10972 |
| chr10_-_128180265 | 0.08 |
ENSMUST00000099139.1
|
Rbms2
|
RNA binding motif, single stranded interacting protein 2 |
| chr15_+_37425554 | 0.08 |
ENSMUST00000022897.1
|
4930447A16Rik
|
RIKEN cDNA 4930447A16 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa10
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.9 | 6.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.6 | 5.5 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.5 | 4.8 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.4 | 2.2 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.4 | 1.5 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.3 | 2.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.3 | 1.8 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.3 | 1.2 | GO:1903383 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.3 | 1.3 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.3 | 2.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 0.7 | GO:0002358 | B cell homeostatic proliferation(GO:0002358) |
| 0.2 | 1.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 | 3.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 1.4 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.2 | 1.3 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.2 | 1.0 | GO:0009446 | spermidine biosynthetic process(GO:0008295) putrescine biosynthetic process(GO:0009446) |
| 0.2 | 0.9 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 | 0.7 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 | 0.6 | GO:1902047 | polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 3.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 4.1 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.1 | 0.7 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.8 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 1.1 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 1.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.5 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 1.4 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 0.7 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 4.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 1.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 5.8 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 0.7 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.8 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 1.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.7 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 0.7 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.4 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.4 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 2.9 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 0.9 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.2 | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination(GO:0010705) |
| 0.1 | 2.0 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 | 0.3 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 1.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.7 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.2 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.0 | 2.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.0 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.6 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 2.2 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.8 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.3 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.8 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.5 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.0 | 0.2 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.5 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.6 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.9 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 2.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 1.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.5 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 2.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 1.9 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 1.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 3.5 | GO:0006869 | lipid transport(GO:0006869) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 1.4 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.2 | 1.5 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 1.3 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.2 | 2.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 6.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 1.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.3 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 0.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 2.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.7 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.1 | 0.5 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.3 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 3.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 6.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.6 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.5 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 1.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 2.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 1.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 3.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 6.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 5.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) outer acrosomal membrane(GO:0002081) |
| 0.0 | 1.9 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.1 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.8 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 1.0 | 6.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.9 | 3.7 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.8 | 4.1 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.8 | 5.5 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.4 | 3.3 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.4 | 2.0 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.4 | 2.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.4 | 2.6 | GO:0005534 | galactose binding(GO:0005534) |
| 0.3 | 2.9 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.3 | 1.4 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.3 | 3.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 1.4 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.2 | 0.9 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.2 | 8.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 0.7 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.2 | 1.2 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.2 | 1.3 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 0.6 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 4.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 1.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 1.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.5 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 3.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.4 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.7 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.7 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 1.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.7 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 1.0 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 2.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.3 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 2.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 1.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 2.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 2.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.5 | GO:0023023 | MHC protein complex binding(GO:0023023) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 1.6 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 1.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 2.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.6 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 2.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 2.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 2.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 0.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 1.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 6.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 1.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 2.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 0.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 4.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 3.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |