Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Hdx
Z-value: 3.68
Transcription factors associated with Hdx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hdx
|
ENSMUSG00000034551.6 | highly divergent homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hdx | mm10_v2_chrX_-_111697069_111697127 | -0.56 | 4.0e-04 | Click! |
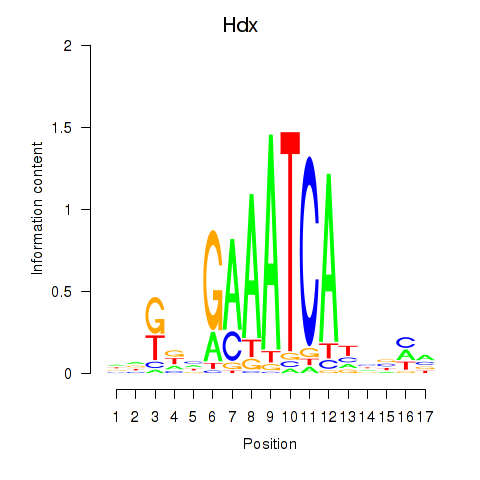
Activity profile of Hdx motif
Sorted Z-values of Hdx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_62150810 | 43.62 |
ENSMUST00000077719.3
|
Mup21
|
major urinary protein 21 |
| chr4_-_60662358 | 42.26 |
ENSMUST00000084544.4
ENSMUST00000098046.3 |
Mup11
|
major urinary protein 11 |
| chr4_-_60139857 | 39.80 |
ENSMUST00000107490.4
ENSMUST00000074700.2 |
Mup2
|
major urinary protein 2 |
| chr4_-_60582152 | 38.30 |
ENSMUST00000098047.2
|
Mup10
|
major urinary protein 10 |
| chr4_-_61303998 | 37.20 |
ENSMUST00000071005.8
ENSMUST00000075206.5 |
Mup14
|
major urinary protein 14 |
| chr4_-_60421933 | 36.68 |
ENSMUST00000107506.2
ENSMUST00000122381.1 ENSMUST00000118759.1 ENSMUST00000122177.1 |
Mup9
|
major urinary protein 9 |
| chr4_-_61674094 | 36.67 |
ENSMUST00000098040.3
|
Mup18
|
major urinary protein 18 |
| chr4_-_60222580 | 36.12 |
ENSMUST00000095058.4
ENSMUST00000163931.1 |
Mup8
|
major urinary protein 8 |
| chr4_-_61303802 | 35.65 |
ENSMUST00000125461.1
|
Mup14
|
major urinary protein 14 |
| chr15_+_4727175 | 16.12 |
ENSMUST00000162585.1
|
C6
|
complement component 6 |
| chr15_+_4727202 | 15.55 |
ENSMUST00000161997.1
ENSMUST00000022788.8 |
C6
|
complement component 6 |
| chr5_-_87092546 | 15.35 |
ENSMUST00000132667.1
ENSMUST00000145617.1 ENSMUST00000094649.4 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr15_+_4727265 | 13.40 |
ENSMUST00000162350.1
|
C6
|
complement component 6 |
| chr1_+_182564994 | 12.99 |
ENSMUST00000048941.7
ENSMUST00000168514.1 |
Capn8
|
calpain 8 |
| chr14_+_37068042 | 11.17 |
ENSMUST00000057176.3
|
Lrit2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 |
| chr1_-_139781236 | 9.32 |
ENSMUST00000027612.8
ENSMUST00000111989.2 ENSMUST00000111986.2 |
Gm4788
|
predicted gene 4788 |
| chr19_-_46672883 | 9.16 |
ENSMUST00000026012.7
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr6_-_3968357 | 8.08 |
ENSMUST00000031674.8
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr15_+_10177623 | 7.90 |
ENSMUST00000124470.1
|
Prlr
|
prolactin receptor |
| chr17_-_34743849 | 7.62 |
ENSMUST00000069507.8
|
C4b
|
complement component 4B (Chido blood group) |
| chr12_-_98577940 | 7.49 |
ENSMUST00000110113.1
|
Kcnk10
|
potassium channel, subfamily K, member 10 |
| chr10_+_62071014 | 7.38 |
ENSMUST00000053865.5
|
Gm5424
|
predicted gene 5424 |
| chr17_+_3397189 | 7.14 |
ENSMUST00000072156.6
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr5_-_86518578 | 6.46 |
ENSMUST00000134179.1
|
Tmprss11g
|
transmembrane protease, serine 11g |
| chr5_-_87535113 | 6.37 |
ENSMUST00000120150.1
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr2_+_69380431 | 6.03 |
ENSMUST00000063690.3
|
Dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr1_+_88070765 | 5.87 |
ENSMUST00000073772.4
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr15_+_10249560 | 5.68 |
ENSMUST00000134410.1
|
Prlr
|
prolactin receptor |
| chr1_+_13668739 | 5.38 |
ENSMUST00000088542.3
|
Xkr9
|
X Kell blood group precursor related family member 9 homolog |
| chr13_-_56548534 | 5.17 |
ENSMUST00000062806.4
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr7_+_13398115 | 5.01 |
ENSMUST00000005791.7
|
Cabp5
|
calcium binding protein 5 |
| chr1_-_140183404 | 5.00 |
ENSMUST00000066859.6
ENSMUST00000111976.2 |
Cfh
|
complement component factor h |
| chr2_+_120977017 | 4.99 |
ENSMUST00000067582.7
|
Tmem62
|
transmembrane protein 62 |
| chr1_-_184811170 | 4.75 |
ENSMUST00000048462.6
ENSMUST00000110992.2 |
Marc1
|
mitochondrial amidoxime reducing component 1 |
| chr17_+_31433054 | 4.75 |
ENSMUST00000136384.1
|
Pde9a
|
phosphodiesterase 9A |
| chr16_+_23107413 | 4.56 |
ENSMUST00000023599.6
ENSMUST00000168891.1 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr3_+_62419668 | 4.50 |
ENSMUST00000161057.1
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr15_-_99087817 | 4.40 |
ENSMUST00000064462.3
|
C1ql4
|
complement component 1, q subcomponent-like 4 |
| chr9_-_45204083 | 4.34 |
ENSMUST00000034599.8
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr9_+_70207342 | 4.12 |
ENSMUST00000034745.7
|
Myo1e
|
myosin IE |
| chr17_-_35895920 | 3.94 |
ENSMUST00000059740.8
|
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chrX_-_21089229 | 3.75 |
ENSMUST00000040667.6
|
Zfp300
|
zinc finger protein 300 |
| chr1_-_140183283 | 3.67 |
ENSMUST00000111977.1
|
Cfh
|
complement component factor h |
| chr14_-_45477856 | 3.62 |
ENSMUST00000141424.1
|
Fermt2
|
fermitin family homolog 2 (Drosophila) |
| chr8_-_84773381 | 3.58 |
ENSMUST00000109764.1
|
Nfix
|
nuclear factor I/X |
| chr4_+_97777606 | 3.57 |
ENSMUST00000075448.6
ENSMUST00000092532.6 |
Nfia
|
nuclear factor I/A |
| chr11_+_97801917 | 3.42 |
ENSMUST00000127033.2
|
Lasp1
|
LIM and SH3 protein 1 |
| chr10_+_34483400 | 3.35 |
ENSMUST00000019913.7
ENSMUST00000170771.1 |
Frk
|
fyn-related kinase |
| chr2_+_116900152 | 3.32 |
ENSMUST00000126467.1
ENSMUST00000128305.1 ENSMUST00000155323.1 |
D330050G23Rik
|
RIKEN cDNA D330050G23 gene |
| chr9_-_71168657 | 3.31 |
ENSMUST00000113570.1
|
Aqp9
|
aquaporin 9 |
| chr14_+_52810934 | 3.30 |
ENSMUST00000103646.3
|
Trav10d
|
T cell receptor alpha variable 10D |
| chr6_+_54267131 | 3.23 |
ENSMUST00000114402.2
|
Chn2
|
chimerin (chimaerin) 2 |
| chr4_+_89137122 | 3.15 |
ENSMUST00000058030.7
|
Mtap
|
methylthioadenosine phosphorylase |
| chr17_-_45686899 | 3.14 |
ENSMUST00000156254.1
|
Tmem63b
|
transmembrane protein 63b |
| chr2_-_126783416 | 3.09 |
ENSMUST00000130356.1
ENSMUST00000028842.2 |
Usp50
|
ubiquitin specific peptidase 50 |
| chr8_-_109579056 | 3.08 |
ENSMUST00000074898.6
|
Hp
|
haptoglobin |
| chr6_-_4086914 | 3.05 |
ENSMUST00000049166.4
|
Bet1
|
blocked early in transport 1 homolog (S. cerevisiae) |
| chr10_-_77799133 | 3.04 |
ENSMUST00000178581.1
|
Gm19668
|
predicted gene, 19668 |
| chr2_-_90022064 | 3.04 |
ENSMUST00000099758.1
|
Olfr1264
|
olfactory receptor 1264 |
| chr9_+_3023547 | 3.03 |
ENSMUST00000099046.3
|
Gm10718
|
predicted gene 10718 |
| chr4_+_145696161 | 3.03 |
ENSMUST00000180014.1
|
Gm13242
|
predicted gene 13242 |
| chr10_-_127121125 | 3.01 |
ENSMUST00000164259.1
ENSMUST00000080975.4 |
Os9
|
amplified in osteosarcoma |
| chr9_+_3025417 | 2.96 |
ENSMUST00000075573.6
|
Gm10717
|
predicted gene 10717 |
| chr11_+_98446826 | 2.57 |
ENSMUST00000019456.4
|
Grb7
|
growth factor receptor bound protein 7 |
| chr11_-_69369377 | 2.56 |
ENSMUST00000092971.6
ENSMUST00000108661.1 |
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr14_-_19418930 | 2.53 |
ENSMUST00000177817.1
|
Gm21738
|
predicted gene, 21738 |
| chr6_-_130193112 | 2.45 |
ENSMUST00000112032.1
ENSMUST00000071554.2 |
Klra9
|
killer cell lectin-like receptor subfamily A, member 9 |
| chr18_-_35498856 | 2.41 |
ENSMUST00000025215.8
|
Sil1
|
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
| chr9_+_3013140 | 2.34 |
ENSMUST00000143083.2
|
Gm10721
|
predicted gene 10721 |
| chr10_+_127420334 | 2.30 |
ENSMUST00000171434.1
|
R3hdm2
|
R3H domain containing 2 |
| chr11_-_69685537 | 2.27 |
ENSMUST00000018896.7
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr6_+_34709610 | 2.22 |
ENSMUST00000031775.6
|
Cald1
|
caldesmon 1 |
| chr3_-_146770603 | 2.22 |
ENSMUST00000106138.1
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr13_+_24327415 | 2.17 |
ENSMUST00000167746.1
|
Cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr11_+_115765420 | 2.15 |
ENSMUST00000093912.4
ENSMUST00000136720.1 ENSMUST00000103034.3 ENSMUST00000141871.1 |
2310067B10Rik
|
RIKEN cDNA 2310067B10 gene |
| chr2_-_121807024 | 2.13 |
ENSMUST00000138157.1
|
Frmd5
|
FERM domain containing 5 |
| chr7_-_7337493 | 2.12 |
ENSMUST00000072475.6
ENSMUST00000174368.1 |
Vmn2r30
|
vomeronasal 2, receptor 30 |
| chr2_-_121806988 | 2.10 |
ENSMUST00000110592.1
|
Frmd5
|
FERM domain containing 5 |
| chr9_+_3004457 | 2.08 |
ENSMUST00000178348.1
|
Gm11168
|
predicted gene 11168 |
| chr11_+_117232254 | 2.08 |
ENSMUST00000106354.2
|
Sept9
|
septin 9 |
| chr3_+_116008220 | 2.08 |
ENSMUST00000106502.1
|
Extl2
|
exostoses (multiple)-like 2 |
| chr5_-_146963742 | 2.02 |
ENSMUST00000125217.1
ENSMUST00000110564.1 ENSMUST00000066675.3 ENSMUST00000016654.2 ENSMUST00000110566.1 ENSMUST00000140526.1 |
Mtif3
|
mitochondrial translational initiation factor 3 |
| chrX_+_134187492 | 2.02 |
ENSMUST00000064476.4
|
Arl13a
|
ADP-ribosylation factor-like 13A |
| chr7_-_89980935 | 2.01 |
ENSMUST00000107234.1
|
Eed
|
embryonic ectoderm development |
| chr2_-_155729359 | 1.99 |
ENSMUST00000040833.4
|
Edem2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr19_-_32061438 | 1.94 |
ENSMUST00000096119.4
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr1_+_180101144 | 1.91 |
ENSMUST00000133890.1
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr13_+_60601921 | 1.90 |
ENSMUST00000077453.5
|
Dapk1
|
death associated protein kinase 1 |
| chr2_-_37359274 | 1.90 |
ENSMUST00000009174.8
|
Pdcl
|
phosducin-like |
| chr17_+_33432890 | 1.87 |
ENSMUST00000174088.2
|
Actl9
|
actin-like 9 |
| chr3_-_34081256 | 1.81 |
ENSMUST00000117223.1
ENSMUST00000120805.1 ENSMUST00000011029.5 ENSMUST00000108195.3 |
Dnajc19
|
DnaJ (Hsp40) homolog, subfamily C, member 19 |
| chr7_-_7247328 | 1.78 |
ENSMUST00000170922.1
|
Vmn2r29
|
vomeronasal 2, receptor 29 |
| chr9_+_3037111 | 1.72 |
ENSMUST00000177969.1
|
Gm10715
|
predicted gene 10715 |
| chr13_+_41249841 | 1.70 |
ENSMUST00000165561.2
|
Smim13
|
small integral membrane protein 13 |
| chr5_+_107597696 | 1.67 |
ENSMUST00000112651.1
ENSMUST00000112654.1 ENSMUST00000065422.5 |
Rpap2
|
RNA polymerase II associated protein 2 |
| chr3_+_121671317 | 1.66 |
ENSMUST00000098646.3
|
4930432M17Rik
|
RIKEN cDNA 4930432M17 gene |
| chr3_-_146770218 | 1.65 |
ENSMUST00000106137.1
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr7_+_81057589 | 1.64 |
ENSMUST00000107348.1
|
Alpk3
|
alpha-kinase 3 |
| chr11_+_102285161 | 1.59 |
ENSMUST00000156326.1
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr3_-_154330543 | 1.54 |
ENSMUST00000184966.1
ENSMUST00000177846.2 |
Lhx8
|
LIM homeobox protein 8 |
| chr2_+_97467657 | 1.52 |
ENSMUST00000059049.7
|
Lrrc4c
|
leucine rich repeat containing 4C |
| chr14_-_68655804 | 1.52 |
ENSMUST00000111072.1
ENSMUST00000022642.5 |
Adam28
|
a disintegrin and metallopeptidase domain 28 |
| chr4_-_43031429 | 1.49 |
ENSMUST00000136326.1
|
Stoml2
|
stomatin (Epb7.2)-like 2 |
| chr5_+_107597760 | 1.48 |
ENSMUST00000112655.1
|
Rpap2
|
RNA polymerase II associated protein 2 |
| chr19_+_12460749 | 1.47 |
ENSMUST00000081035.7
|
Mpeg1
|
macrophage expressed gene 1 |
| chr9_+_3034599 | 1.42 |
ENSMUST00000178641.1
|
Gm17535
|
predicted gene, 17535 |
| chr9_-_83254460 | 1.37 |
ENSMUST00000184080.1
ENSMUST00000184100.1 |
RP23-341H6.1
|
RP23-341H6.1 |
| chr7_+_131174400 | 1.37 |
ENSMUST00000050586.5
|
5430419D17Rik
|
RIKEN cDNA 5430419D17 gene |
| chr11_-_100050542 | 1.36 |
ENSMUST00000007318.1
|
Krt31
|
keratin 31 |
| chr14_+_62663665 | 1.35 |
ENSMUST00000171692.1
|
Serpine3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
| chrX_+_143518671 | 1.31 |
ENSMUST00000134402.1
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr18_-_31949571 | 1.31 |
ENSMUST00000064016.5
|
Gpr17
|
G protein-coupled receptor 17 |
| chr12_-_81568474 | 1.29 |
ENSMUST00000008582.3
|
Adam21
|
a disintegrin and metallopeptidase domain 21 |
| chr4_-_134000857 | 1.28 |
ENSMUST00000105887.1
ENSMUST00000012262.5 ENSMUST00000144668.1 ENSMUST00000105889.3 |
Dhdds
|
dehydrodolichyl diphosphate synthase |
| chr4_+_3938904 | 1.19 |
ENSMUST00000120732.1
ENSMUST00000041122.4 ENSMUST00000121651.1 ENSMUST00000121210.1 ENSMUST00000119307.1 ENSMUST00000123769.1 |
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr4_-_43031370 | 1.18 |
ENSMUST00000138030.1
|
Stoml2
|
stomatin (Epb7.2)-like 2 |
| chr4_+_124880899 | 1.18 |
ENSMUST00000059343.6
|
Epha10
|
Eph receptor A10 |
| chr7_-_9841325 | 1.18 |
ENSMUST00000170131.1
|
Vmn2r47
|
vomeronasal 2, receptor 47 |
| chr16_-_37539781 | 1.14 |
ENSMUST00000023525.8
|
Gtf2e1
|
general transcription factor II E, polypeptide 1 (alpha subunit) |
| chr11_-_48946148 | 1.12 |
ENSMUST00000104958.1
|
Psme2b
|
protease (prosome, macropain) activator subunit 2B |
| chr5_+_32863700 | 1.12 |
ENSMUST00000149350.1
ENSMUST00000118698.1 ENSMUST00000150130.1 ENSMUST00000087897.4 ENSMUST00000119705.1 ENSMUST00000125574.1 ENSMUST00000049780.6 |
Depdc5
|
DEP domain containing 5 |
| chr1_+_87327008 | 1.09 |
ENSMUST00000172794.1
ENSMUST00000164992.2 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr7_-_9572288 | 1.09 |
ENSMUST00000174433.1
|
Gm10302
|
predicted gene 10302 |
| chr1_+_130865669 | 1.09 |
ENSMUST00000038829.5
|
Faim3
|
Fas apoptotic inhibitory molecule 3 |
| chr5_-_109097864 | 1.04 |
ENSMUST00000095922.3
|
Vmn2r12
|
vomeronasal 2, receptor 12 |
| chr11_-_35798884 | 1.01 |
ENSMUST00000160726.2
|
Fbll1
|
fibrillarin-like 1 |
| chr13_+_60602182 | 1.01 |
ENSMUST00000044083.7
|
Dapk1
|
death associated protein kinase 1 |
| chr12_-_27160311 | 1.00 |
ENSMUST00000182473.1
ENSMUST00000177636.1 ENSMUST00000183238.1 |
Gm9866
|
predicted gene 9866 |
| chr7_-_128596278 | 0.99 |
ENSMUST00000179317.1
|
Gm7258
|
predicted gene 7258 |
| chr8_-_41016295 | 0.98 |
ENSMUST00000131965.1
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr10_+_85928491 | 0.98 |
ENSMUST00000170396.1
|
Ascl4
|
achaete-scute complex homolog 4 (Drosophila) |
| chr5_+_107437908 | 0.97 |
ENSMUST00000094541.2
|
Btbd8
|
BTB (POZ) domain containing 8 |
| chr9_+_3005125 | 0.97 |
ENSMUST00000179881.1
|
Gm11168
|
predicted gene 11168 |
| chr3_-_129755305 | 0.96 |
ENSMUST00000029653.2
|
Egf
|
epidermal growth factor |
| chr6_-_143947061 | 0.94 |
ENSMUST00000124233.1
|
Sox5
|
SRY-box containing gene 5 |
| chrX_+_143518576 | 0.94 |
ENSMUST00000033640.7
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr12_-_27160498 | 0.92 |
ENSMUST00000182592.1
|
Gm9866
|
predicted gene 9866 |
| chr14_-_36968679 | 0.91 |
ENSMUST00000067700.6
|
Ccser2
|
coiled-coil serine rich 2 |
| chr14_-_72602945 | 0.91 |
ENSMUST00000162825.1
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr16_+_35938470 | 0.87 |
ENSMUST00000114878.1
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr11_+_4257557 | 0.86 |
ENSMUST00000066283.5
|
Lif
|
leukemia inhibitory factor |
| chr9_+_3036877 | 0.86 |
ENSMUST00000155807.2
|
Gm10715
|
predicted gene 10715 |
| chr16_-_50330987 | 0.85 |
ENSMUST00000114488.1
|
Bbx
|
bobby sox homolog (Drosophila) |
| chr14_+_53845234 | 0.83 |
ENSMUST00000103674.4
|
Trav19
|
T cell receptor alpha variable 19 |
| chr4_+_43406435 | 0.83 |
ENSMUST00000098106.2
ENSMUST00000139198.1 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr10_-_130429612 | 0.83 |
ENSMUST00000171811.2
|
Vmn2r85
|
vomeronasal 2, receptor 85 |
| chr9_+_94669876 | 0.82 |
ENSMUST00000033463.9
|
Slc9a9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
| chr6_-_57535422 | 0.81 |
ENSMUST00000042766.3
|
Ppm1k
|
protein phosphatase 1K (PP2C domain containing) |
| chr1_+_180109192 | 0.81 |
ENSMUST00000143176.1
ENSMUST00000135056.1 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr6_-_143947092 | 0.81 |
ENSMUST00000144289.1
ENSMUST00000111748.1 |
Sox5
|
SRY-box containing gene 5 |
| chr9_+_3000922 | 0.80 |
ENSMUST00000151376.2
|
Gm10722
|
predicted gene 10722 |
| chr11_-_98193260 | 0.79 |
ENSMUST00000092735.5
ENSMUST00000107545.2 |
Med1
|
mediator complex subunit 1 |
| chr7_-_42933789 | 0.79 |
ENSMUST00000163803.1
|
Vmn2r63
|
vomeronasal 2, receptor 63 |
| chr9_+_3015654 | 0.77 |
ENSMUST00000099050.3
|
Gm10720
|
predicted gene 10720 |
| chr5_-_26004798 | 0.77 |
ENSMUST00000168875.1
|
Gm1979
|
predicted gene 1979 |
| chr8_+_109493982 | 0.74 |
ENSMUST00000034162.6
|
Pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr5_-_26089291 | 0.73 |
ENSMUST00000094946.4
|
Gm10471
|
predicted gene 10471 |
| chr4_-_97584605 | 0.69 |
ENSMUST00000107067.1
|
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr13_-_67729109 | 0.68 |
ENSMUST00000073157.6
|
Zfp71-rs1
|
zinc finger protein 71, related sequence |
| chr5_-_25926215 | 0.68 |
ENSMUST00000167847.2
|
Gm21655
|
predicted gene, 21655 |
| chr13_+_43370710 | 0.66 |
ENSMUST00000066804.4
|
Sirt5
|
sirtuin 5 |
| chr13_-_66355385 | 0.66 |
ENSMUST00000099416.3
|
Vmn2r-ps104
|
vomeronasal 2, receptor, pseudogene 104 |
| chr14_+_13454010 | 0.66 |
ENSMUST00000112656.2
|
Synpr
|
synaptoporin |
| chr5_-_107597577 | 0.66 |
ENSMUST00000100949.3
ENSMUST00000078021.6 |
Glmn
|
glomulin, FKBP associated protein |
| chr5_+_26257691 | 0.64 |
ENSMUST00000074148.6
|
Gm7361
|
predicted gene 7361 |
| chr18_+_77375065 | 0.64 |
ENSMUST00000123410.1
|
Loxhd1
|
lipoxygenase homology domains 1 |
| chr16_+_14705832 | 0.64 |
ENSMUST00000023356.6
|
Snai2
|
snail homolog 2 (Drosophila) |
| chr7_+_13398150 | 0.61 |
ENSMUST00000117400.1
|
Cabp5
|
calcium binding protein 5 |
| chr5_-_25954344 | 0.61 |
ENSMUST00000162387.4
|
Gm21671
|
predicted gene, 21671 |
| chr16_-_50432340 | 0.61 |
ENSMUST00000066037.6
ENSMUST00000089399.4 ENSMUST00000089404.3 ENSMUST00000114477.1 ENSMUST00000138166.1 |
Bbx
|
bobby sox homolog (Drosophila) |
| chr8_-_85025268 | 0.59 |
ENSMUST00000064314.8
|
Asna1
|
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
| chr16_+_22951072 | 0.58 |
ENSMUST00000023590.8
|
Hrg
|
histidine-rich glycoprotein |
| chr9_+_45055166 | 0.57 |
ENSMUST00000114664.1
ENSMUST00000093856.3 |
Mpzl3
|
myelin protein zero-like 3 |
| chr1_+_87327044 | 0.57 |
ENSMUST00000173173.1
|
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr9_+_87729285 | 0.55 |
ENSMUST00000181359.1
|
D030062O11Rik
|
RIKEN cDNA D030062O11 gene |
| chr5_-_26105362 | 0.54 |
ENSMUST00000063524.2
|
5031410I06Rik
|
RIKEN cDNA 5031410I06 gene |
| chr5_-_15714236 | 0.54 |
ENSMUST00000095005.5
ENSMUST00000179506.1 |
Speer4c
|
spermatogenesis associated glutamate (E)-rich protein 4c |
| chr5_-_109192107 | 0.52 |
ENSMUST00000053253.8
|
Vmn2r13
|
vomeronasal 2, receptor 13 |
| chr5_-_26121421 | 0.52 |
ENSMUST00000088236.3
|
Gm10220
|
predicted gene 10220 |
| chr5_-_14938429 | 0.51 |
ENSMUST00000159973.2
|
Speer4e
|
spermatogenesis associated glutamate (E)-rich protein 4e |
| chr5_-_26039506 | 0.50 |
ENSMUST00000079447.2
|
Speer4a
|
spermatogenesis associated glutamate (E)-rich protein 4a |
| chr6_-_130699016 | 0.50 |
ENSMUST00000182643.1
|
Gm26919
|
predicted gene, 26919 |
| chr2_-_41789078 | 0.47 |
ENSMUST00000167270.2
|
Lrp1b
|
low density lipoprotein-related protein 1B (deleted in tumors) |
| chr15_+_102954427 | 0.46 |
ENSMUST00000001701.3
|
Hoxc11
|
homeobox C11 |
| chr10_-_130497379 | 0.45 |
ENSMUST00000164227.2
|
Vmn2r87
|
vomeronasal 2, receptor 87 |
| chr5_-_14978935 | 0.43 |
ENSMUST00000096953.4
|
Gm10354
|
predicted gene 10354 |
| chr3_-_145099024 | 0.43 |
ENSMUST00000040465.6
|
Clca5
|
chloride channel calcium activated 5 |
| chr5_+_15619064 | 0.42 |
ENSMUST00000095006.2
|
Speer4d
|
spermatogenesis associated glutamate (E)-rich protein 4d |
| chr18_-_39489157 | 0.42 |
ENSMUST00000131885.1
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr1_+_87326997 | 0.40 |
ENSMUST00000027475.8
|
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr7_-_41872670 | 0.40 |
ENSMUST00000171671.1
|
Vmn2r58
|
vomeronasal 2, receptor 58 |
| chr4_-_97584612 | 0.39 |
ENSMUST00000107068.2
|
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr7_-_8260599 | 0.39 |
ENSMUST00000066317.4
|
Vmn2r43
|
vomeronasal 2, receptor 43 |
| chr9_+_45055211 | 0.39 |
ENSMUST00000114663.2
|
Mpzl3
|
myelin protein zero-like 3 |
| chr16_-_4624984 | 0.38 |
ENSMUST00000014445.6
|
Pam16
|
presequence translocase-asssociated motor 16 homolog (S. cerevisiae) |
| chr11_+_95666957 | 0.36 |
ENSMUST00000125172.1
ENSMUST00000036374.5 |
Phb
|
prohibitin |
| chr7_-_44257204 | 0.35 |
ENSMUST00000012921.7
|
Acpt
|
acid phosphatase, testicular |
| chr5_-_129879038 | 0.35 |
ENSMUST00000026617.6
|
Phkg1
|
phosphorylase kinase gamma 1 |
| chr5_-_86518562 | 0.34 |
ENSMUST00000140095.1
|
Tmprss11g
|
transmembrane protease, serine 11g |
| chr7_-_9953585 | 0.33 |
ENSMUST00000165611.1
|
Vmn2r48
|
vomeronasal 2, receptor 48 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hdx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.0 | 45.1 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 3.8 | 42.3 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 2.3 | 13.6 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 1.1 | 6.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 1.0 | 3.1 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 1.0 | 8.7 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.9 | 2.7 | GO:0090297 | positive regulation of mitochondrial DNA replication(GO:0090297) regulation of cardiolipin metabolic process(GO:1900208) positive regulation of cardiolipin metabolic process(GO:1900210) stress-induced mitochondrial fusion(GO:1990046) |
| 0.9 | 6.0 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.8 | 3.3 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.8 | 3.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.8 | 2.3 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.7 | 4.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.7 | 2.0 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.6 | 1.9 | GO:1902605 | heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.6 | 4.8 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.5 | 2.2 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.5 | 9.2 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.5 | 7.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.5 | 5.9 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.4 | 3.0 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.4 | 2.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.4 | 0.4 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.4 | 3.9 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.3 | 3.1 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.3 | 0.6 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.3 | 2.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.3 | 2.0 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.3 | 4.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 1.0 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.2 | 3.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 1.0 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.2 | 3.6 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 1.7 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.2 | 0.6 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.2 | 4.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.2 | 3.6 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 2.9 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.2 | 0.8 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.9 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) leukemia inhibitory factor signaling pathway(GO:0048861) positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.1 | 1.3 | GO:0016093 | polyprenol metabolic process(GO:0016093) |
| 0.1 | 7.6 | GO:0006956 | complement activation(GO:0006956) |
| 0.1 | 3.6 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 2.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 4.4 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 1.3 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 2.7 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 2.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 13.0 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 0.4 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 | 2.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 2.2 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.1 | 0.2 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.1 | 1.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.8 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.9 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.1 | 3.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.7 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.0 | 0.5 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.2 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 8.1 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 2.2 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.9 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 2.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.3 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 1.0 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 1.8 | GO:0048806 | genitalia development(GO:0048806) |
| 0.0 | 5.2 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 1.5 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 1.6 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 3.1 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 2.6 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 1.1 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 1.5 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 4.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.2 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.0 | 1.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.8 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 45.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.7 | 4.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.6 | 3.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 3.0 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.3 | 3.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 1.5 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.3 | 7.6 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.2 | 2.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 1.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 2.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 0.6 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.2 | 1.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 2.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 2.1 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 2.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 2.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 3.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 8.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 3.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 4.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0031464 | Cul2-RING ubiquitin ligase complex(GO:0031462) Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 2.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 6.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 3.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.4 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 4.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 4.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.2 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 2.7 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 7.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 13.6 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 1.6 | 4.8 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 1.3 | 9.0 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 1.3 | 6.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 1.1 | 3.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 1.0 | 6.0 | GO:0047035 | alcohol dehydrogenase (NAD) activity(GO:0004022) testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 1.0 | 7.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.7 | 2.2 | GO:0030338 | CMP-N-acetylneuraminate monooxygenase activity(GO:0030338) |
| 0.7 | 2.1 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.7 | 3.3 | GO:0015254 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.6 | 9.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.6 | 21.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.5 | 7.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.4 | 3.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.4 | 3.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 3.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 1.9 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.3 | 2.7 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 2.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 1.9 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 2.0 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 0.9 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.2 | 4.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.2 | 0.8 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.2 | 4.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 0.5 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.2 | 1.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 2.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 5.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 2.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 2.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.4 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 1.3 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 4.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 2.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.3 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 10.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.0 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 3.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.2 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.1 | 0.3 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 2.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 2.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.5 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 1.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.7 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 4.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 3.1 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 4.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 2.9 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 14.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 3.9 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 3.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 2.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 3.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 5.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 5.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 3.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 3.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 53.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.7 | 9.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.7 | 13.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.6 | 7.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.5 | 6.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.5 | 5.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 3.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 2.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 3.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 2.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 4.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 3.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 4.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 2.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.0 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 4.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 2.3 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 2.1 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |