Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
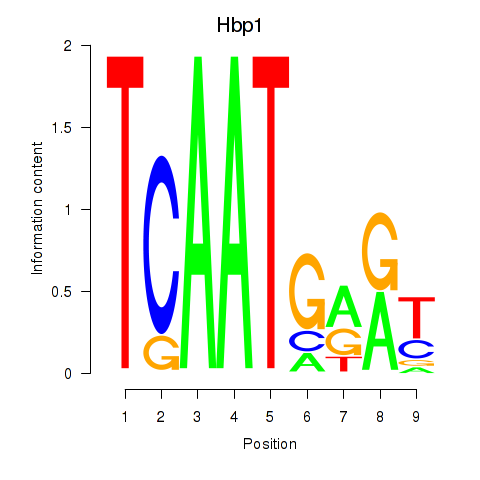
Results for Hbp1
Z-value: 1.94
Transcription factors associated with Hbp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hbp1
|
ENSMUSG00000002996.11 | high mobility group box transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hbp1 | mm10_v2_chr12_-_31950535_31950560 | 0.21 | 2.1e-01 | Click! |
Activity profile of Hbp1 motif
Sorted Z-values of Hbp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_127898515 | 13.34 |
ENSMUST00000047134.7
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr5_-_86926521 | 12.21 |
ENSMUST00000031183.2
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr19_+_39992424 | 11.12 |
ENSMUST00000049178.2
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr19_-_8131982 | 10.28 |
ENSMUST00000065651.4
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr12_-_104153846 | 10.04 |
ENSMUST00000085050.3
|
Serpina3c
|
serine (or cysteine) peptidase inhibitor, clade A, member 3C |
| chr19_+_39007019 | 9.41 |
ENSMUST00000025966.4
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr17_-_32917320 | 8.58 |
ENSMUST00000179434.1
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr19_+_39287074 | 8.36 |
ENSMUST00000003137.8
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr5_-_87337165 | 7.93 |
ENSMUST00000031195.2
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr4_-_115496129 | 7.37 |
ENSMUST00000030487.2
|
Cyp4a14
|
cytochrome P450, family 4, subfamily a, polypeptide 14 |
| chr1_+_88070765 | 7.31 |
ENSMUST00000073772.4
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr1_+_88095054 | 6.90 |
ENSMUST00000150634.1
ENSMUST00000058237.7 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr8_+_67490758 | 6.82 |
ENSMUST00000026677.3
|
Nat1
|
N-acetyl transferase 1 |
| chr4_+_115299046 | 6.60 |
ENSMUST00000084343.3
|
Cyp4a12a
|
cytochrome P450, family 4, subfamily a, polypeptide 12a |
| chr13_+_4436094 | 6.50 |
ENSMUST00000156277.1
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr19_-_8405060 | 5.98 |
ENSMUST00000064507.5
ENSMUST00000120540.1 ENSMUST00000096269.4 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr8_-_123754138 | 5.84 |
ENSMUST00000181805.1
|
4732419C18Rik
|
RIKEN cDNA 4732419C18 gene |
| chr1_+_78511865 | 5.65 |
ENSMUST00000012331.6
|
Mogat1
|
monoacylglycerol O-acyltransferase 1 |
| chr3_-_86999284 | 5.53 |
ENSMUST00000063869.5
ENSMUST00000029717.2 |
Cd1d1
|
CD1d1 antigen |
| chr3_+_94372794 | 5.53 |
ENSMUST00000029795.3
|
Rorc
|
RAR-related orphan receptor gamma |
| chr7_+_44207307 | 5.45 |
ENSMUST00000077354.4
|
Klk1b4
|
kallikrein 1-related pepidase b4 |
| chr9_-_48605147 | 5.36 |
ENSMUST00000034808.5
ENSMUST00000119426.1 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr19_+_12633507 | 5.30 |
ENSMUST00000119960.1
|
Glyat
|
glycine-N-acyltransferase |
| chr19_+_30232921 | 5.07 |
ENSMUST00000025797.5
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chr8_+_60506109 | 5.02 |
ENSMUST00000079472.2
|
Aadat
|
aminoadipate aminotransferase |
| chr3_+_138277489 | 4.95 |
ENSMUST00000004232.9
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr3_-_85722474 | 4.94 |
ENSMUST00000119077.1
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr19_+_12633303 | 4.86 |
ENSMUST00000044976.5
|
Glyat
|
glycine-N-acyltransferase |
| chr6_-_138079916 | 4.57 |
ENSMUST00000171804.1
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr19_-_4498574 | 4.53 |
ENSMUST00000048482.6
|
2010003K11Rik
|
RIKEN cDNA 2010003K11 gene |
| chr5_-_87140318 | 4.37 |
ENSMUST00000067790.6
ENSMUST00000113327.1 |
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr1_+_67123015 | 4.37 |
ENSMUST00000027144.7
|
Cps1
|
carbamoyl-phosphate synthetase 1 |
| chr1_+_88211956 | 4.23 |
ENSMUST00000073049.6
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr13_+_23807027 | 4.21 |
ENSMUST00000006786.4
ENSMUST00000099697.2 |
Slc17a2
|
solute carrier family 17 (sodium phosphate), member 2 |
| chr2_+_58754910 | 4.20 |
ENSMUST00000059102.6
|
Upp2
|
uridine phosphorylase 2 |
| chr7_+_87246649 | 4.17 |
ENSMUST00000068829.5
ENSMUST00000032781.7 |
Nox4
|
NADPH oxidase 4 |
| chr13_-_19307551 | 4.12 |
ENSMUST00000103561.1
|
Tcrg-C2
|
T-cell receptor gamma, constant 2 |
| chr2_+_173153048 | 4.10 |
ENSMUST00000029017.5
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr6_+_41302265 | 4.05 |
ENSMUST00000031913.4
|
Try4
|
trypsin 4 |
| chr6_-_85933379 | 4.04 |
ENSMUST00000162660.1
|
Nat8b
|
N-acetyltransferase 8B |
| chr6_-_41035501 | 3.91 |
ENSMUST00000031931.5
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr5_-_145879857 | 3.89 |
ENSMUST00000035918.7
|
Cyp3a11
|
cytochrome P450, family 3, subfamily a, polypeptide 11 |
| chr1_+_74332596 | 3.86 |
ENSMUST00000087225.5
|
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr10_+_93488766 | 3.85 |
ENSMUST00000129421.1
|
Hal
|
histidine ammonia lyase |
| chr4_-_104876383 | 3.79 |
ENSMUST00000064873.8
ENSMUST00000106808.3 ENSMUST00000048947.8 |
C8a
|
complement component 8, alpha polypeptide |
| chr5_+_90561102 | 3.57 |
ENSMUST00000094615.4
|
5830473C10Rik
|
RIKEN cDNA 5830473C10 gene |
| chr8_-_41215146 | 3.56 |
ENSMUST00000034003.4
|
Fgl1
|
fibrinogen-like protein 1 |
| chr7_-_68275098 | 3.51 |
ENSMUST00000135564.1
|
Gm16157
|
predicted gene 16157 |
| chr10_-_81291227 | 3.51 |
ENSMUST00000045744.6
|
Tjp3
|
tight junction protein 3 |
| chr14_+_55561060 | 3.48 |
ENSMUST00000117701.1
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr7_-_132576372 | 3.43 |
ENSMUST00000084500.6
|
Oat
|
ornithine aminotransferase |
| chr5_-_147322435 | 3.34 |
ENSMUST00000100433.4
|
Urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5) decarboxylase |
| chr7_-_3249711 | 3.34 |
ENSMUST00000108653.2
|
Nlrp12
|
NLR family, pyrin domain containing 12 |
| chr18_+_12643329 | 3.32 |
ENSMUST00000025294.7
|
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr11_-_113710017 | 3.29 |
ENSMUST00000018871.1
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr14_-_30943275 | 3.28 |
ENSMUST00000006704.8
ENSMUST00000163118.1 |
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr3_+_146597077 | 3.24 |
ENSMUST00000029837.7
ENSMUST00000121133.1 |
Uox
|
urate oxidase |
| chr1_+_78511805 | 3.24 |
ENSMUST00000152111.1
|
Mogat1
|
monoacylglycerol O-acyltransferase 1 |
| chr3_-_98509967 | 3.21 |
ENSMUST00000179429.1
|
Hsd3b4
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 4 |
| chr8_+_104733997 | 3.21 |
ENSMUST00000034346.8
ENSMUST00000164182.2 |
Ces2a
|
carboxylesterase 2A |
| chr19_+_38481057 | 3.21 |
ENSMUST00000182481.1
|
Plce1
|
phospholipase C, epsilon 1 |
| chr7_+_44590886 | 3.18 |
ENSMUST00000107906.3
|
Kcnc3
|
potassium voltage gated channel, Shaw-related subfamily, member 3 |
| chr8_-_117673682 | 3.15 |
ENSMUST00000173522.1
ENSMUST00000174450.1 |
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr19_+_42036025 | 3.12 |
ENSMUST00000026172.2
|
Ankrd2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr15_-_98677451 | 3.08 |
ENSMUST00000120997.1
ENSMUST00000109149.2 ENSMUST00000003451.4 |
Rnd1
|
Rho family GTPase 1 |
| chr2_-_86347764 | 3.02 |
ENSMUST00000099894.2
|
Olfr1055
|
olfactory receptor 1055 |
| chr1_+_88055377 | 3.01 |
ENSMUST00000138182.1
ENSMUST00000113142.3 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr4_-_59960659 | 3.00 |
ENSMUST00000075973.2
|
Mup4
|
major urinary protein 4 |
| chr7_+_51880312 | 3.00 |
ENSMUST00000145049.1
|
Gas2
|
growth arrest specific 2 |
| chr1_+_88055467 | 2.93 |
ENSMUST00000173325.1
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr1_+_88087802 | 2.93 |
ENSMUST00000113139.1
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr18_-_38866702 | 2.91 |
ENSMUST00000115582.1
|
Fgf1
|
fibroblast growth factor 1 |
| chr19_-_40187277 | 2.85 |
ENSMUST00000051846.6
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr2_-_25501717 | 2.84 |
ENSMUST00000015227.3
|
C8g
|
complement component 8, gamma polypeptide |
| chr8_+_45658666 | 2.79 |
ENSMUST00000134675.1
ENSMUST00000139869.1 ENSMUST00000126067.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr7_+_100022099 | 2.78 |
ENSMUST00000144808.1
|
Chrdl2
|
chordin-like 2 |
| chr5_+_115466234 | 2.75 |
ENSMUST00000145785.1
ENSMUST00000031495.4 ENSMUST00000112071.1 ENSMUST00000125568.1 |
Pla2g1b
|
phospholipase A2, group IB, pancreas |
| chr5_-_87591582 | 2.74 |
ENSMUST00000031201.7
|
Sult1e1
|
sulfotransferase family 1E, member 1 |
| chr11_-_43901187 | 2.72 |
ENSMUST00000067258.2
ENSMUST00000139906.1 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr1_+_88138364 | 2.70 |
ENSMUST00000014263.4
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr6_-_85762480 | 2.68 |
ENSMUST00000168531.1
|
Cml3
|
camello-like 3 |
| chr3_-_113574758 | 2.68 |
ENSMUST00000106540.1
|
Amy1
|
amylase 1, salivary |
| chr7_-_141434402 | 2.67 |
ENSMUST00000136354.1
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr5_-_137921612 | 2.66 |
ENSMUST00000031741.7
|
Cyp3a13
|
cytochrome P450, family 3, subfamily a, polypeptide 13 |
| chr19_-_44029201 | 2.65 |
ENSMUST00000026211.8
|
Cyp2c44
|
cytochrome P450, family 2, subfamily c, polypeptide 44 |
| chr17_+_79614900 | 2.64 |
ENSMUST00000040368.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr2_-_52335134 | 2.61 |
ENSMUST00000075301.3
|
Neb
|
nebulin |
| chr4_-_130279205 | 2.59 |
ENSMUST00000120126.2
|
Serinc2
|
serine incorporator 2 |
| chr8_+_45658273 | 2.56 |
ENSMUST00000153798.1
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr1_+_88200601 | 2.54 |
ENSMUST00000049289.8
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr3_-_113324052 | 2.52 |
ENSMUST00000179314.1
|
Amy2a3
|
amylase 2a3 |
| chr9_-_51328898 | 2.50 |
ENSMUST00000039959.4
|
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr19_+_37685581 | 2.47 |
ENSMUST00000073391.4
|
Cyp26c1
|
cytochrome P450, family 26, subfamily c, polypeptide 1 |
| chrX_+_10252305 | 2.47 |
ENSMUST00000049910.6
|
Otc
|
ornithine transcarbamylase |
| chr11_-_26210553 | 2.45 |
ENSMUST00000101447.3
|
5730522E02Rik
|
RIKEN cDNA 5730522E02 gene |
| chrX_-_147429189 | 2.44 |
ENSMUST00000033646.2
|
Il13ra2
|
interleukin 13 receptor, alpha 2 |
| chrX_+_10252361 | 2.43 |
ENSMUST00000115528.2
|
Otc
|
ornithine transcarbamylase |
| chr16_+_22951072 | 2.43 |
ENSMUST00000023590.8
|
Hrg
|
histidine-rich glycoprotein |
| chr5_-_86518578 | 2.40 |
ENSMUST00000134179.1
|
Tmprss11g
|
transmembrane protease, serine 11g |
| chr7_-_139582790 | 2.38 |
ENSMUST00000106095.2
|
Nkx6-2
|
NK6 homeobox 2 |
| chr1_-_140183404 | 2.34 |
ENSMUST00000066859.6
ENSMUST00000111976.2 |
Cfh
|
complement component factor h |
| chr8_+_45628176 | 2.34 |
ENSMUST00000130850.1
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr11_+_83709015 | 2.32 |
ENSMUST00000001009.7
|
Wfdc18
|
WAP four-disulfide core domain 18 |
| chr2_-_64975762 | 2.32 |
ENSMUST00000156765.1
|
Grb14
|
growth factor receptor bound protein 14 |
| chr8_-_94696223 | 2.28 |
ENSMUST00000034227.4
|
Pllp
|
plasma membrane proteolipid |
| chr16_-_10543028 | 2.25 |
ENSMUST00000184863.1
ENSMUST00000038281.5 |
Dexi
|
dexamethasone-induced transcript |
| chr16_-_19200350 | 2.22 |
ENSMUST00000103749.2
|
Iglc2
|
immunoglobulin lambda constant 2 |
| chr1_-_180245927 | 2.12 |
ENSMUST00000010753.7
|
Psen2
|
presenilin 2 |
| chr9_+_57697612 | 2.11 |
ENSMUST00000034865.4
|
Cyp1a1
|
cytochrome P450, family 1, subfamily a, polypeptide 1 |
| chr11_+_75468040 | 2.10 |
ENSMUST00000043598.7
ENSMUST00000108435.1 |
Tlcd2
|
TLC domain containing 2 |
| chr4_+_124657646 | 2.10 |
ENSMUST00000053491.7
|
Pou3f1
|
POU domain, class 3, transcription factor 1 |
| chr13_+_4049001 | 2.10 |
ENSMUST00000118717.2
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr2_-_67194695 | 2.06 |
ENSMUST00000147939.1
|
Gm13598
|
predicted gene 13598 |
| chr6_+_72636244 | 2.05 |
ENSMUST00000101278.2
|
Gm15401
|
predicted gene 15401 |
| chr1_+_131797381 | 2.03 |
ENSMUST00000112393.2
ENSMUST00000048660.5 |
Pm20d1
|
peptidase M20 domain containing 1 |
| chr1_-_153851189 | 2.03 |
ENSMUST00000059607.6
|
5830403L16Rik
|
RIKEN cDNA 5830403L16 gene |
| chr5_-_87424201 | 2.03 |
ENSMUST00000072818.5
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr8_+_45627946 | 2.02 |
ENSMUST00000145458.1
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr13_+_65512678 | 2.01 |
ENSMUST00000081471.2
|
Gm10139
|
predicted gene 10139 |
| chr8_-_3467617 | 2.00 |
ENSMUST00000111081.3
ENSMUST00000118194.1 ENSMUST00000004686.6 |
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr3_-_113291449 | 1.99 |
ENSMUST00000179568.1
|
Amy2a4
|
amylase 2a4 |
| chr1_+_93135244 | 1.99 |
ENSMUST00000027491.5
|
Agxt
|
alanine-glyoxylate aminotransferase |
| chr3_-_113258837 | 1.97 |
ENSMUST00000098673.3
|
Amy2a5
|
amylase 2a5 |
| chr1_-_140183283 | 1.97 |
ENSMUST00000111977.1
|
Cfh
|
complement component factor h |
| chr11_-_110251736 | 1.96 |
ENSMUST00000044003.7
|
Abca6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr8_-_86580664 | 1.96 |
ENSMUST00000131423.1
ENSMUST00000152438.1 |
Abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr1_+_21240597 | 1.96 |
ENSMUST00000121676.1
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr1_+_21240581 | 1.94 |
ENSMUST00000027067.8
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr2_-_35100677 | 1.92 |
ENSMUST00000045776.4
ENSMUST00000113050.3 |
AI182371
|
expressed sequence AI182371 |
| chr5_-_120711927 | 1.92 |
ENSMUST00000031607.6
|
Dtx1
|
deltex 1 homolog (Drosophila) |
| chr5_+_90518932 | 1.92 |
ENSMUST00000113179.2
ENSMUST00000128740.1 |
Afm
|
afamin |
| chr19_-_30175414 | 1.91 |
ENSMUST00000025778.7
|
Gldc
|
glycine decarboxylase |
| chr17_-_31636631 | 1.90 |
ENSMUST00000135425.1
ENSMUST00000151718.1 ENSMUST00000155814.1 |
Cbs
|
cystathionine beta-synthase |
| chr8_+_45627709 | 1.90 |
ENSMUST00000134321.1
ENSMUST00000135336.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr3_-_113532288 | 1.90 |
ENSMUST00000132353.1
|
Amy2a1
|
amylase 2a1 |
| chr4_-_118134869 | 1.90 |
ENSMUST00000097912.1
ENSMUST00000030263.2 ENSMUST00000106410.1 |
St3gal3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| chr19_-_4439388 | 1.86 |
ENSMUST00000117462.1
ENSMUST00000048197.3 |
Rhod
|
ras homolog gene family, member D |
| chr8_+_12395287 | 1.85 |
ENSMUST00000180353.1
|
Sox1
|
SRY-box containing gene 1 |
| chr17_+_56005672 | 1.84 |
ENSMUST00000133998.1
|
Mpnd
|
MPN domain containing |
| chr15_+_76268076 | 1.84 |
ENSMUST00000074173.3
|
Spatc1
|
spermatogenesis and centriole associated 1 |
| chr2_+_177508570 | 1.80 |
ENSMUST00000108940.2
|
Gm14403
|
predicted gene 14403 |
| chr18_-_3281036 | 1.80 |
ENSMUST00000049942.6
ENSMUST00000139537.1 ENSMUST00000124747.1 |
Crem
|
cAMP responsive element modulator |
| chr8_-_119635553 | 1.79 |
ENSMUST00000061828.3
|
Kcng4
|
potassium voltage-gated channel, subfamily G, member 4 |
| chr3_-_88296838 | 1.77 |
ENSMUST00000010682.3
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr4_+_80910646 | 1.76 |
ENSMUST00000055922.3
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr3_+_60031754 | 1.76 |
ENSMUST00000029325.3
|
Aadac
|
arylacetamide deacetylase (esterase) |
| chr4_+_135920731 | 1.75 |
ENSMUST00000030434.4
|
Fuca1
|
fucosidase, alpha-L- 1, tissue |
| chr14_-_75787031 | 1.75 |
ENSMUST00000022580.6
|
Slc25a30
|
solute carrier family 25, member 30 |
| chr13_+_4059565 | 1.75 |
ENSMUST00000041768.6
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr10_+_57784914 | 1.74 |
ENSMUST00000165013.1
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr17_+_29268788 | 1.73 |
ENSMUST00000064709.5
ENSMUST00000120346.1 |
BC004004
|
cDNA sequence BC004004 |
| chrM_+_14138 | 1.72 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr12_-_80968075 | 1.70 |
ENSMUST00000095572.4
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr8_+_45658731 | 1.70 |
ENSMUST00000143820.1
ENSMUST00000132139.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr9_-_76323559 | 1.67 |
ENSMUST00000063140.8
|
Hcrtr2
|
hypocretin (orexin) receptor 2 |
| chr17_+_36958623 | 1.67 |
ENSMUST00000173814.1
|
Znrd1as
|
Znrd1 antisense |
| chr3_-_88296979 | 1.67 |
ENSMUST00000107556.3
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr3_-_146596588 | 1.66 |
ENSMUST00000029836.4
|
Dnase2b
|
deoxyribonuclease II beta |
| chr17_-_73950172 | 1.65 |
ENSMUST00000024866.4
|
Xdh
|
xanthine dehydrogenase |
| chr11_-_77519186 | 1.65 |
ENSMUST00000100807.2
|
Gm10392
|
predicted gene 10392 |
| chr16_+_37580137 | 1.65 |
ENSMUST00000160847.1
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr16_+_56477838 | 1.64 |
ENSMUST00000048471.7
ENSMUST00000096013.3 ENSMUST00000096012.3 ENSMUST00000171000.1 |
Abi3bp
|
ABI gene family, member 3 (NESH) binding protein |
| chr1_-_180245757 | 1.63 |
ENSMUST00000111104.1
|
Psen2
|
presenilin 2 |
| chr6_-_124741374 | 1.63 |
ENSMUST00000004389.5
|
Grcc10
|
gene rich cluster, C10 gene |
| chr4_+_144893127 | 1.62 |
ENSMUST00000142808.1
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr4_+_100095791 | 1.60 |
ENSMUST00000039630.5
|
Ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr2_+_32609043 | 1.59 |
ENSMUST00000128811.1
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr4_-_130275523 | 1.59 |
ENSMUST00000146478.1
|
Serinc2
|
serine incorporator 2 |
| chr13_+_93674403 | 1.58 |
ENSMUST00000048001.6
|
Dmgdh
|
dimethylglycine dehydrogenase precursor |
| chr13_+_4233730 | 1.58 |
ENSMUST00000081326.6
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr15_-_39857459 | 1.56 |
ENSMUST00000022915.3
ENSMUST00000110306.1 |
Dpys
|
dihydropyrimidinase |
| chr10_+_29143996 | 1.55 |
ENSMUST00000092629.2
|
Soga3
|
SOGA family member 3 |
| chr3_+_121953213 | 1.53 |
ENSMUST00000037958.7
ENSMUST00000128366.1 |
Arhgap29
|
Rho GTPase activating protein 29 |
| chr13_-_56548534 | 1.53 |
ENSMUST00000062806.4
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr3_+_89459325 | 1.52 |
ENSMUST00000107410.1
|
Pmvk
|
phosphomevalonate kinase |
| chr7_+_16452779 | 1.52 |
ENSMUST00000019302.8
|
Tmem160
|
transmembrane protein 160 |
| chr9_-_106891965 | 1.52 |
ENSMUST00000159283.1
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr11_-_50931612 | 1.52 |
ENSMUST00000109124.3
|
Zfp354b
|
zinc finger protein 354B |
| chr5_-_86906937 | 1.51 |
ENSMUST00000031181.9
ENSMUST00000113333.1 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr11_-_115062177 | 1.51 |
ENSMUST00000062787.7
|
Cd300e
|
CD300e antigen |
| chr18_-_35627223 | 1.51 |
ENSMUST00000025212.5
|
Slc23a1
|
solute carrier family 23 (nucleobase transporters), member 1 |
| chr4_-_141723401 | 1.49 |
ENSMUST00000102484.4
ENSMUST00000177592.1 |
Ddi2
Rsc1a1
|
DNA-damage inducible protein 2 regulatory solute carrier protein, family 1, member 1 |
| chr17_-_34862122 | 1.47 |
ENSMUST00000154526.1
|
Cfb
|
complement factor B |
| chr14_-_30353468 | 1.47 |
ENSMUST00000112249.1
|
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr10_-_81545175 | 1.47 |
ENSMUST00000043604.5
|
Gna11
|
guanine nucleotide binding protein, alpha 11 |
| chr11_+_77518566 | 1.46 |
ENSMUST00000147386.1
|
Abhd15
|
abhydrolase domain containing 15 |
| chrM_-_14060 | 1.45 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr10_+_57784859 | 1.45 |
ENSMUST00000020024.5
|
Fabp7
|
fatty acid binding protein 7, brain |
| chrX_-_100594860 | 1.45 |
ENSMUST00000053373.1
|
P2ry4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr8_-_3694167 | 1.44 |
ENSMUST00000005678.4
|
Fcer2a
|
Fc receptor, IgE, low affinity II, alpha polypeptide |
| chr11_-_115187827 | 1.43 |
ENSMUST00000103041.1
|
Nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr6_+_78405148 | 1.43 |
ENSMUST00000023906.2
|
Reg2
|
regenerating islet-derived 2 |
| chr13_-_66852017 | 1.43 |
ENSMUST00000059329.6
|
Gm17449
|
predicted gene, 17449 |
| chr19_+_20405280 | 1.42 |
ENSMUST00000179640.1
|
1500015L24Rik
|
RIKEN cDNA 1500015L24 gene |
| chr17_-_85090204 | 1.41 |
ENSMUST00000072406.3
ENSMUST00000171795.1 |
Prepl
|
prolyl endopeptidase-like |
| chr12_+_78691516 | 1.41 |
ENSMUST00000077968.4
|
Fam71d
|
family with sequence similarity 71, member D |
| chr5_-_86676346 | 1.41 |
ENSMUST00000038448.6
|
Tmprss11bnl
|
transmembrane protease, serine 11b N terminal like |
| chr17_-_34862473 | 1.40 |
ENSMUST00000025229.4
ENSMUST00000176203.2 ENSMUST00000128767.1 |
Cfb
|
complement factor B |
| chr9_+_74861888 | 1.40 |
ENSMUST00000056006.9
|
Onecut1
|
one cut domain, family member 1 |
| chr9_-_106891870 | 1.38 |
ENSMUST00000160503.1
ENSMUST00000159620.2 ENSMUST00000160978.1 |
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hbp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.4 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 3.0 | 9.0 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 2.2 | 8.9 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 2.2 | 6.5 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 2.1 | 21.4 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 2.0 | 7.9 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 1.8 | 5.5 | GO:0048007 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 1.6 | 4.9 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 1.4 | 4.1 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 1.3 | 3.9 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 1.3 | 7.7 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 1.2 | 16.3 | GO:0015747 | urate transport(GO:0015747) |
| 1.1 | 3.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 1.1 | 35.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.0 | 2.9 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.9 | 2.8 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.9 | 2.8 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.9 | 2.7 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.9 | 4.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.9 | 13.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.8 | 2.4 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.8 | 3.8 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.7 | 2.0 | GO:0019448 | cysteine catabolic process(GO:0009093) glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-cysteine catabolic process(GO:0019448) L-alanine catabolic process(GO:0042853) L-cysteine metabolic process(GO:0046439) |
| 0.6 | 1.9 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.6 | 1.9 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 0.6 | 2.5 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.6 | 2.5 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.6 | 4.2 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.6 | 1.8 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 0.6 | 2.9 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.5 | 1.6 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.5 | 5.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.5 | 5.5 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.5 | 3.4 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.5 | 5.3 | GO:0070189 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.5 | 0.9 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.5 | 3.7 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.4 | 1.6 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.4 | 1.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.4 | 3.9 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.4 | 1.1 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.4 | 1.1 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.4 | 1.5 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.4 | 2.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.4 | 1.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.4 | 1.1 | GO:1990705 | intrahepatic bile duct development(GO:0035622) cholangiocyte proliferation(GO:1990705) |
| 0.4 | 4.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.3 | 1.7 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.3 | 2.4 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.3 | 2.0 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.3 | 3.7 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.3 | 1.3 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.3 | 2.0 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.3 | 1.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.3 | 0.3 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.3 | 1.2 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.3 | 7.6 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.3 | 4.2 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.3 | 2.3 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.3 | 3.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.3 | 1.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.3 | 3.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 1.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.3 | 1.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.3 | 1.1 | GO:1902477 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.3 | 1.3 | GO:0006041 | glucosamine metabolic process(GO:0006041) UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.3 | 1.0 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.3 | 3.8 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 1.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.2 | 1.2 | GO:0060295 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.2 | 1.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 1.9 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.2 | 1.6 | GO:1902548 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.2 | 0.7 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.2 | 0.7 | GO:1903048 | regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.2 | 0.7 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.2 | 0.4 | GO:0002362 | CD4-positive, CD25-positive, alpha-beta regulatory T cell lineage commitment(GO:0002362) |
| 0.2 | 1.8 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.2 | 0.7 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.2 | 1.7 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 | 1.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.2 | 1.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 0.6 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 1.4 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.2 | 0.8 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.2 | 2.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 3.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 1.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 1.8 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.2 | 0.7 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.2 | 1.5 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 10.9 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 0.7 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.2 | 5.5 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.2 | 3.2 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.2 | 0.5 | GO:2000832 | negative regulation of steroid hormone secretion(GO:2000832) |
| 0.2 | 0.5 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.2 | 0.8 | GO:0009446 | spermidine biosynthetic process(GO:0008295) putrescine biosynthetic process(GO:0009446) |
| 0.2 | 0.8 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.2 | 0.3 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 0.4 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.1 | 0.4 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 0.9 | GO:0042997 | protein depalmitoylation(GO:0002084) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.4 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.9 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.1 | 0.6 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.7 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 | 0.4 | GO:1990869 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) negative regulation of lymphangiogenesis(GO:1901491) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.1 | 0.5 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.7 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.7 | GO:0060066 | oviduct development(GO:0060066) |
| 0.1 | 1.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 2.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.6 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.4 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 1.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 1.2 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.5 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.1 | 4.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 0.5 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.4 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.1 | 1.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 1.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 1.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.7 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.3 | GO:0060618 | nipple development(GO:0060618) |
| 0.1 | 0.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.3 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 0.7 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.1 | 0.4 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.1 | 2.5 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 0.7 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 3.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 1.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 2.1 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 1.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.4 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.3 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 1.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.8 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 0.7 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 1.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.2 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.1 | 0.6 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.1 | 0.5 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 0.6 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 0.5 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.3 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.1 | 0.5 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 2.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.2 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.1 | 1.0 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 1.3 | GO:0097502 | mannosylation(GO:0097502) |
| 0.1 | 0.3 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.1 | 0.2 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.1 | 0.7 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.5 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.0 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.1 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.1 | 1.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.5 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 2.2 | GO:0045581 | negative regulation of T cell differentiation(GO:0045581) |
| 0.1 | 1.4 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 0.6 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.1 | 0.9 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 1.7 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 7.9 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 0.8 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.5 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 1.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.3 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 1.4 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.1 | 0.8 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.1 | 0.7 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.1 | 0.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 1.5 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 0.3 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.8 | GO:0044036 | cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.1 | 0.2 | GO:0042697 | menstrual cycle phase(GO:0022601) menopause(GO:0042697) |
| 0.1 | 0.3 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 5.4 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.5 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.4 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 3.6 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.2 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 1.0 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.8 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.8 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.4 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.9 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.3 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.9 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.0 | GO:0046514 | ceramide catabolic process(GO:0046514) |
| 0.0 | 0.4 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 1.9 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.3 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.9 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 2.8 | GO:0032835 | glomerulus development(GO:0032835) |
| 0.0 | 1.7 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 1.1 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 1.6 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 1.2 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:1902856 | microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.0 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.0 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.2 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.8 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.2 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.7 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 | 1.0 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.8 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.2 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.5 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 2.1 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.7 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.9 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.1 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) positive regulation of tooth mineralization(GO:0070172) regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.9 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.3 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.1 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.0 | 0.1 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 1.4 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.5 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.3 | GO:0021930 | cell proliferation in hindbrain(GO:0021534) cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.0 | 0.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 1.1 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.3 | GO:0045019 | negative regulation of nitric oxide biosynthetic process(GO:0045019) negative regulation of nitric oxide metabolic process(GO:1904406) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 1.3 | GO:0061045 | negative regulation of wound healing(GO:0061045) |
| 0.0 | 1.2 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 1.2 | GO:0010659 | cardiac muscle cell apoptotic process(GO:0010659) |
| 0.0 | 0.6 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.5 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 3.3 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.0 | 3.6 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.0 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.9 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.3 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.3 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.2 | GO:0060770 | epithelial cell proliferation involved in prostate gland development(GO:0060767) regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.5 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 3.3 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.5 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.7 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.6 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:1903020 | positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.7 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.5 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.5 | GO:0001832 | blastocyst growth(GO:0001832) |
| 0.0 | 0.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.9 | GO:0090109 | regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.1 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.7 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) regulation of nonmotile primary cilium assembly(GO:1902855) |
| 0.0 | 0.3 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.7 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 2.3 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.6 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.2 | GO:0007616 | long-term memory(GO:0007616) positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.6 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.4 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.0 | 0.2 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.3 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.3 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 2.0 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.2 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 0.3 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.7 | 5.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.6 | 6.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.5 | 2.5 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.4 | 1.6 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.4 | 4.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.4 | 1.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.3 | 1.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.3 | 4.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 1.4 | GO:0030891 | VCB complex(GO:0030891) |
| 0.2 | 0.7 | GO:0036019 | endolysosome(GO:0036019) |
| 0.2 | 3.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 0.5 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 1.7 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.2 | 1.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 3.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.9 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.1 | 1.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 21.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 6.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.4 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 1.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.5 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.5 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 2.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 4.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.3 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 1.0 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 1.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 1.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.6 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 1.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.3 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 3.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 2.0 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 13.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 1.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.8 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.1 | 0.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.3 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 3.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 2.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 94.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 2.7 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 1.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 2.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 8.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 3.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 2.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 3.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 7.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 6.7 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.1 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.4 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 1.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 1.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 2.8 | 14.0 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 2.5 | 10.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 2.5 | 4.9 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 2.4 | 9.4 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 2.2 | 8.9 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 2.2 | 6.5 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 1.7 | 6.8 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 1.7 | 5.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 1.6 | 60.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 1.5 | 9.0 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 1.5 | 4.4 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 1.3 | 4.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 1.3 | 9.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 1.2 | 17.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 1.1 | 6.7 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 1.1 | 5.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.0 | 23.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.8 | 4.2 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.8 | 2.5 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.8 | 15.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.7 | 5.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.7 | 5.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.7 | 4.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.7 | 2.7 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.6 | 1.9 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.6 | 4.3 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.6 | 1.8 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.6 | 2.2 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.6 | 1.7 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.5 | 3.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.5 | 2.0 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.5 | 1.5 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.5 | 1.5 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.5 | 1.9 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.5 | 3.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.5 | 3.6 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.5 | 6.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.4 | 1.3 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.4 | 3.9 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.4 | 1.7 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.4 | 1.6 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.4 | 1.2 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.4 | 10.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.4 | 7.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.4 | 4.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.4 | 1.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.4 | 1.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.4 | 1.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.3 | 2.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 2.7 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.3 | 1.6 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.3 | 1.2 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.3 | 1.2 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.3 | 1.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 1.9 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 1.7 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 0.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 2.7 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 0.6 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.2 | 1.5 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.2 | 0.7 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.2 | 4.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 1.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 1.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 2.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 1.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 0.5 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.2 | 0.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 0.8 | GO:0008384 | NF-kappaB-inducing kinase activity(GO:0004704) IkappaB kinase activity(GO:0008384) |
| 0.2 | 0.5 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 0.6 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.2 | 0.8 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.1 | 0.9 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 2.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.9 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 2.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 3.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 2.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.5 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.6 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 1.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.5 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 2.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 3.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.5 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 2.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 1.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 2.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 1.1 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.3 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.7 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 1.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.5 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 12.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.3 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) |
| 0.1 | 0.7 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 2.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.3 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.3 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 2.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 3.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 7.5 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 1.9 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.1 | 0.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.9 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 2.5 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.1 | 3.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 1.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.5 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.2 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.1 | 0.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.2 | GO:0003681 | bent DNA binding(GO:0003681) |
| 0.1 | 0.7 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.2 | GO:0019962 | interferon receptor activity(GO:0004904) type I interferon receptor activity(GO:0004905) type I interferon binding(GO:0019962) |
| 0.1 | 2.0 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 0.2 | GO:0051381 | histamine binding(GO:0051381) |
| 0.1 | 0.2 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 0.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 1.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 2.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 2.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 1.0 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.8 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.8 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.7 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 7.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 1.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.5 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 1.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 2.8 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 3.6 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 2.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 1.1 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 2.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 4.3 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.0 | 0.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 1.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.4 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 3.0 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 1.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.1 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 2.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.0 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 2.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 4.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.4 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 0.9 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 1.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 0.9 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 5.1 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 1.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 4.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 4.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 4.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 3.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 2.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.4 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.9 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 3.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.3 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 4.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 14.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.8 | 6.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.7 | 2.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.6 | 13.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 5.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 4.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 22.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.3 | 4.8 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.3 | 5.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 1.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 1.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 3.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 2.9 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 1.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 3.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 3.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 2.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 6.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 3.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 16.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 1.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 3.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 0.5 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 1.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 5.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.8 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.9 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 3.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 4.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.1 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME MEIOSIS | Genes involved in Meiosis |