Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
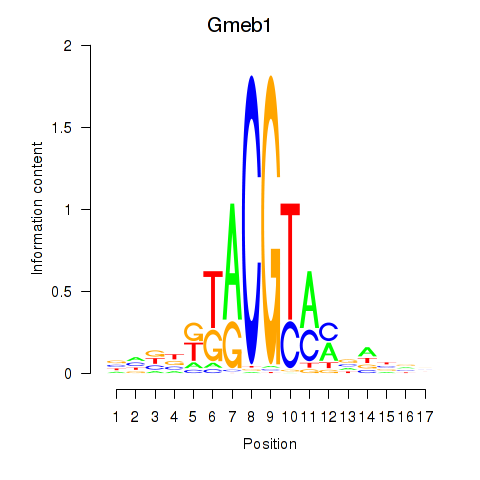
Results for Gmeb1
Z-value: 1.58
Transcription factors associated with Gmeb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gmeb1
|
ENSMUSG00000028901.7 | glucocorticoid modulatory element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gmeb1 | mm10_v2_chr4_-_132261521_132261572 | 0.87 | 4.1e-12 | Click! |
Activity profile of Gmeb1 motif
Sorted Z-values of Gmeb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_23139064 | 9.58 |
ENSMUST00000033947.8
|
Ank1
|
ankyrin 1, erythroid |
| chr8_+_23139030 | 8.57 |
ENSMUST00000121075.1
|
Ank1
|
ankyrin 1, erythroid |
| chr6_+_113531675 | 8.29 |
ENSMUST00000036340.5
ENSMUST00000101051.2 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr3_+_127553462 | 6.25 |
ENSMUST00000043108.4
|
4930422G04Rik
|
RIKEN cDNA 4930422G04 gene |
| chr6_+_90619241 | 6.21 |
ENSMUST00000032177.8
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr2_-_157007015 | 5.82 |
ENSMUST00000146413.1
|
Dsn1
|
DSN1, MIND kinetochore complex component, homolog (S. cerevisiae) |
| chr15_+_99074968 | 5.13 |
ENSMUST00000039665.6
|
Troap
|
trophinin associated protein |
| chr2_-_157007039 | 5.06 |
ENSMUST00000103129.2
ENSMUST00000103130.1 |
Dsn1
|
DSN1, MIND kinetochore complex component, homolog (S. cerevisiae) |
| chr3_+_40800054 | 5.05 |
ENSMUST00000168287.1
|
Plk4
|
polo-like kinase 4 |
| chr1_+_175880775 | 5.03 |
ENSMUST00000039725.6
|
Exo1
|
exonuclease 1 |
| chr3_+_40800013 | 4.74 |
ENSMUST00000026858.5
ENSMUST00000170825.1 |
Plk4
|
polo-like kinase 4 |
| chr19_-_40588374 | 4.51 |
ENSMUST00000175932.1
ENSMUST00000176955.1 ENSMUST00000149476.2 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chrX_-_150814265 | 4.43 |
ENSMUST00000026302.6
ENSMUST00000129768.1 ENSMUST00000112699.2 |
Maged2
|
melanoma antigen, family D, 2 |
| chr19_-_40588453 | 4.39 |
ENSMUST00000025979.6
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr19_-_40588338 | 4.08 |
ENSMUST00000176939.1
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr11_+_69095217 | 3.25 |
ENSMUST00000101004.2
|
Per1
|
period circadian clock 1 |
| chr8_-_123949201 | 2.83 |
ENSMUST00000044795.7
|
Nup133
|
nucleoporin 133 |
| chr8_+_23139157 | 2.78 |
ENSMUST00000174435.1
|
Ank1
|
ankyrin 1, erythroid |
| chr10_-_81378459 | 2.57 |
ENSMUST00000140901.1
|
Fzr1
|
fizzy/cell division cycle 20 related 1 (Drosophila) |
| chr1_-_91459254 | 2.54 |
ENSMUST00000069620.8
|
Per2
|
period circadian clock 2 |
| chr11_+_69914179 | 2.46 |
ENSMUST00000057884.5
|
Gps2
|
G protein pathway suppressor 2 |
| chr16_+_49699198 | 2.34 |
ENSMUST00000046777.4
ENSMUST00000142682.1 |
Ift57
|
intraflagellar transport 57 |
| chr11_-_40733373 | 2.27 |
ENSMUST00000020579.8
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr15_-_58135047 | 1.96 |
ENSMUST00000038194.3
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr19_+_43612299 | 1.78 |
ENSMUST00000057178.9
|
Nkx2-3
|
NK2 homeobox 3 |
| chr2_+_75659253 | 1.74 |
ENSMUST00000111964.1
ENSMUST00000111962.1 ENSMUST00000111961.1 ENSMUST00000164947.2 ENSMUST00000090792.4 |
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr7_-_83884289 | 1.73 |
ENSMUST00000094216.3
|
Mesdc1
|
mesoderm development candidate 1 |
| chr16_+_20696175 | 1.65 |
ENSMUST00000128273.1
|
Fam131a
|
family with sequence similarity 131, member A |
| chr10_+_80798652 | 1.52 |
ENSMUST00000151928.1
|
Sf3a2
|
splicing factor 3a, subunit 2 |
| chr4_-_135353126 | 1.29 |
ENSMUST00000030613.4
ENSMUST00000131373.1 |
Srrm1
|
serine/arginine repetitive matrix 1 |
| chr7_+_35397046 | 1.28 |
ENSMUST00000079414.5
|
Cep89
|
centrosomal protein 89 |
| chr4_-_126202583 | 1.23 |
ENSMUST00000106142.1
ENSMUST00000169403.1 ENSMUST00000130334.1 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr11_+_86544982 | 1.20 |
ENSMUST00000108030.2
ENSMUST00000020821.3 ENSMUST00000069503.6 ENSMUST00000167178.2 |
Tubd1
|
tubulin, delta 1 |
| chr2_+_69723071 | 1.08 |
ENSMUST00000040915.8
|
Ppig
|
peptidyl-prolyl isomerase G (cyclophilin G) |
| chr4_-_135353164 | 1.08 |
ENSMUST00000084846.5
ENSMUST00000136342.2 ENSMUST00000105861.1 |
Srrm1
|
serine/arginine repetitive matrix 1 |
| chr6_-_87672142 | 1.06 |
ENSMUST00000032130.2
ENSMUST00000065997.2 |
Aplf
|
aprataxin and PNKP like factor |
| chr17_+_23679363 | 1.04 |
ENSMUST00000024699.2
|
Cldn6
|
claudin 6 |
| chr14_+_20694956 | 1.00 |
ENSMUST00000048016.1
|
Fut11
|
fucosyltransferase 11 |
| chr9_+_108002501 | 0.97 |
ENSMUST00000035214.4
ENSMUST00000175874.1 |
Ip6k1
|
inositol hexaphosphate kinase 1 |
| chr5_+_93206518 | 0.96 |
ENSMUST00000031330.4
|
2010109A12Rik
|
RIKEN cDNA 2010109A12 gene |
| chr17_+_29549783 | 0.96 |
ENSMUST00000048677.7
|
Tbc1d22b
|
TBC1 domain family, member 22B |
| chr13_+_23574381 | 0.92 |
ENSMUST00000090776.4
|
Hist1h2ad
|
histone cluster 1, H2ad |
| chr2_+_69722797 | 0.87 |
ENSMUST00000090858.3
|
Ppig
|
peptidyl-prolyl isomerase G (cyclophilin G) |
| chr2_-_180334665 | 0.83 |
ENSMUST00000015771.2
|
Gata5
|
GATA binding protein 5 |
| chr12_+_4917376 | 0.77 |
ENSMUST00000045664.5
|
Atad2b
|
ATPase family, AAA domain containing 2B |
| chr3_-_108086590 | 0.70 |
ENSMUST00000102638.1
ENSMUST00000102637.1 |
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr11_-_97782377 | 0.69 |
ENSMUST00000128801.1
|
Rpl23
|
ribosomal protein L23 |
| chr11_+_83302817 | 0.67 |
ENSMUST00000142680.1
|
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr17_+_84511832 | 0.63 |
ENSMUST00000047206.5
|
Plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr19_-_24477356 | 0.62 |
ENSMUST00000099556.1
|
Fam122a
|
family with sequence similarity 122, member A |
| chr8_-_94918012 | 0.60 |
ENSMUST00000077955.5
|
Ccdc102a
|
coiled-coil domain containing 102A |
| chr7_+_5020561 | 0.57 |
ENSMUST00000085427.3
|
Zfp865
|
zinc finger protein 865 |
| chr19_+_46075842 | 0.56 |
ENSMUST00000165017.1
|
Nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr11_-_97782409 | 0.56 |
ENSMUST00000103146.4
|
Rpl23
|
ribosomal protein L23 |
| chr17_+_83350925 | 0.54 |
ENSMUST00000096766.4
ENSMUST00000112363.2 ENSMUST00000049503.8 |
Eml4
|
echinoderm microtubule associated protein like 4 |
| chr7_+_5020376 | 0.53 |
ENSMUST00000076251.4
|
Zfp865
|
zinc finger protein 865 |
| chr4_+_88776922 | 0.51 |
ENSMUST00000179158.1
|
Gm13289
|
predicted gene 13289 |
| chr17_+_85090647 | 0.47 |
ENSMUST00000095188.5
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr4_-_126202757 | 0.46 |
ENSMUST00000080919.5
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr15_-_79546741 | 0.42 |
ENSMUST00000054014.7
|
Ddx17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 |
| chr6_-_128424883 | 0.38 |
ENSMUST00000001559.8
|
Itfg2
|
integrin alpha FG-GAP repeat containing 2 |
| chr3_-_19311269 | 0.33 |
ENSMUST00000099195.3
|
Pde7a
|
phosphodiesterase 7A |
| chr11_+_22990519 | 0.29 |
ENSMUST00000173867.1
ENSMUST00000020562.4 |
Cct4
|
chaperonin containing Tcp1, subunit 4 (delta) |
| chr11_-_74724670 | 0.26 |
ENSMUST00000021091.8
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr3_-_127553233 | 0.26 |
ENSMUST00000029588.5
|
Larp7
|
La ribonucleoprotein domain family, member 7 |
| chr7_-_111082997 | 0.24 |
ENSMUST00000161051.1
ENSMUST00000160132.1 ENSMUST00000106666.3 ENSMUST00000162415.1 |
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chrX_+_112311334 | 0.16 |
ENSMUST00000026599.3
ENSMUST00000113415.1 |
Apool
|
apolipoprotein O-like |
| chr4_+_88779850 | 0.13 |
ENSMUST00000179425.1
|
Gm13272
|
predicted gene 13272 |
| chr17_-_29549588 | 0.10 |
ENSMUST00000114683.2
ENSMUST00000168339.1 |
Tmem217
|
transmembrane protein 217 |
| chr11_+_97315716 | 0.09 |
ENSMUST00000019026.3
ENSMUST00000132168.1 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr10_-_112928974 | 0.01 |
ENSMUST00000099276.2
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr15_-_83724979 | 0.00 |
ENSMUST00000171496.1
ENSMUST00000043634.5 ENSMUST00000076060.5 ENSMUST00000016907.7 |
Scube1
|
signal peptide, CUB domain, EGF-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gmeb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 13.0 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 2.7 | 10.9 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 1.4 | 9.8 | GO:0046601 | positive regulation of centriole replication(GO:0046601) de novo centriole assembly(GO:0098535) |
| 1.2 | 5.8 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.8 | 8.3 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.6 | 4.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.5 | 20.9 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.4 | 5.0 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.3 | 1.8 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.3 | 2.8 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.3 | 2.6 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.2 | 1.2 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.2 | 2.7 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.2 | 1.1 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.1 | 0.4 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 2.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 1.0 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.7 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 1.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.6 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 0.3 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 0.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 2.5 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.8 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.4 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.6 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 1.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.3 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 1.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 1.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.6 | GO:1901880 | negative regulation of protein depolymerization(GO:1901880) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.9 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 1.6 | 9.8 | GO:0098536 | deuterosome(GO:0098536) |
| 0.9 | 20.9 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.3 | 2.8 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 2.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 2.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 1.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 1.5 | GO:0071004 | U2 snRNP(GO:0005686) U2-type prespliceosome(GO:0071004) |
| 0.1 | 1.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 1.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 8.3 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 4.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 11.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 1.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 13.0 | GO:0017084 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 2.1 | 20.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.3 | 5.0 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.4 | 2.6 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.3 | 1.7 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.2 | 7.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 1.0 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.2 | 2.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 1.0 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 1.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 2.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 2.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.7 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 3.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 1.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 8.9 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 1.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 8.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 2.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.2 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 8.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.5 | 13.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.5 | 20.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 2.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 9.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 5.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 11.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.2 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 3.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |