Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
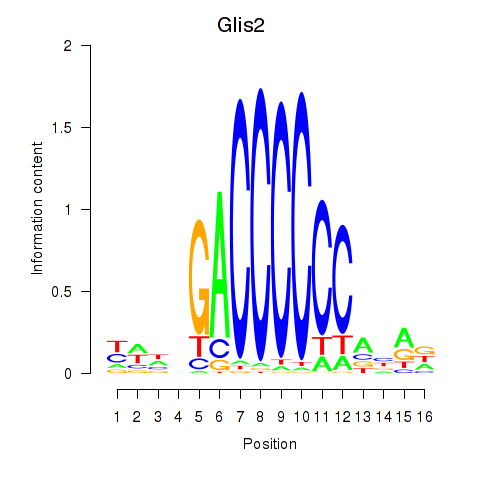
Results for Glis2
Z-value: 1.09
Transcription factors associated with Glis2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Glis2
|
ENSMUSG00000014303.7 | GLIS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Glis2 | mm10_v2_chr16_+_4594683_4594735 | 0.57 | 2.7e-04 | Click! |
Activity profile of Glis2 motif
Sorted Z-values of Glis2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_20269162 | 7.38 |
ENSMUST00000024155.7
|
Kcnk16
|
potassium channel, subfamily K, member 16 |
| chr7_-_103843154 | 6.30 |
ENSMUST00000063957.4
|
Hbb-bh1
|
hemoglobin Z, beta-like embryonic chain |
| chr11_+_87793722 | 5.57 |
ENSMUST00000143021.2
|
Mpo
|
myeloperoxidase |
| chr12_+_109544498 | 5.16 |
ENSMUST00000126289.1
|
Meg3
|
maternally expressed 3 |
| chr2_-_170427828 | 5.06 |
ENSMUST00000013667.2
ENSMUST00000109152.2 ENSMUST00000068137.4 |
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr19_+_7268296 | 4.67 |
ENSMUST00000066646.4
|
Rcor2
|
REST corepressor 2 |
| chr11_+_87793470 | 4.62 |
ENSMUST00000020779.4
|
Mpo
|
myeloperoxidase |
| chr4_-_118620763 | 3.87 |
ENSMUST00000071972.4
|
Wdr65
|
WD repeat domain 65 |
| chr4_+_115059507 | 3.85 |
ENSMUST00000162489.1
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr8_+_72761868 | 3.75 |
ENSMUST00000058099.8
|
F2rl3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr17_-_56830916 | 3.67 |
ENSMUST00000002444.7
ENSMUST00000086801.5 |
Rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr2_+_118111876 | 3.55 |
ENSMUST00000039559.8
|
Thbs1
|
thrombospondin 1 |
| chr10_-_128401218 | 3.55 |
ENSMUST00000042666.5
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr8_+_105690906 | 2.99 |
ENSMUST00000062574.6
|
Rltpr
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing |
| chr7_-_126704736 | 2.67 |
ENSMUST00000131415.1
|
Coro1a
|
coronin, actin binding protein 1A |
| chr7_+_44572370 | 2.64 |
ENSMUST00000002274.8
|
Napsa
|
napsin A aspartic peptidase |
| chr9_+_59680144 | 2.56 |
ENSMUST00000123914.1
|
Gramd2
|
GRAM domain containing 2 |
| chr12_+_109545390 | 2.42 |
ENSMUST00000146701.1
|
Meg3
|
maternally expressed 3 |
| chr2_+_71117923 | 2.37 |
ENSMUST00000028403.2
|
Cybrd1
|
cytochrome b reductase 1 |
| chr7_-_126414855 | 2.21 |
ENSMUST00000032968.5
|
Cd19
|
CD19 antigen |
| chr7_+_43440782 | 2.16 |
ENSMUST00000040227.1
|
Cldnd2
|
claudin domain containing 2 |
| chr7_-_126704816 | 2.09 |
ENSMUST00000032949.7
|
Coro1a
|
coronin, actin binding protein 1A |
| chr4_-_49593875 | 2.04 |
ENSMUST00000151542.1
|
Tmem246
|
transmembrane protein 246 |
| chr17_-_46629420 | 1.91 |
ENSMUST00000044442.8
|
Ptk7
|
PTK7 protein tyrosine kinase 7 |
| chr14_+_55854115 | 1.91 |
ENSMUST00000168479.1
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr1_+_86303221 | 1.89 |
ENSMUST00000113306.2
|
B3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr2_-_152830615 | 1.78 |
ENSMUST00000146380.1
ENSMUST00000134902.1 ENSMUST00000134357.1 ENSMUST00000109820.3 |
Bcl2l1
|
BCL2-like 1 |
| chr11_+_72042455 | 1.77 |
ENSMUST00000021164.3
|
Fam64a
|
family with sequence similarity 64, member A |
| chr10_-_127341583 | 1.72 |
ENSMUST00000026474.3
|
Gli1
|
GLI-Kruppel family member GLI1 |
| chr3_+_130180882 | 1.71 |
ENSMUST00000106353.1
ENSMUST00000080335.4 |
Col25a1
|
collagen, type XXV, alpha 1 |
| chr9_+_107975529 | 1.57 |
ENSMUST00000035216.4
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr10_-_127189981 | 1.57 |
ENSMUST00000019611.7
|
Arhgef25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr2_-_164743182 | 1.57 |
ENSMUST00000103096.3
|
Wfdc3
|
WAP four-disulfide core domain 3 |
| chr8_+_70501116 | 1.48 |
ENSMUST00000127983.1
|
Crlf1
|
cytokine receptor-like factor 1 |
| chr7_+_127746775 | 1.47 |
ENSMUST00000033081.7
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr3_+_105870858 | 1.46 |
ENSMUST00000164730.1
|
Adora3
|
adenosine A3 receptor |
| chr5_-_137212389 | 1.41 |
ENSMUST00000179412.1
|
A630081J09Rik
|
RIKEN cDNA A630081J09 gene |
| chr2_-_152831112 | 1.38 |
ENSMUST00000128172.1
|
Bcl2l1
|
BCL2-like 1 |
| chr4_+_104367549 | 1.33 |
ENSMUST00000106830.2
|
Dab1
|
disabled 1 |
| chr17_-_45549655 | 1.31 |
ENSMUST00000180252.1
|
Tmem151b
|
transmembrane protein 151B |
| chr4_-_133498538 | 1.30 |
ENSMUST00000125541.1
|
Trnp1
|
TMF1-regulated nuclear protein 1 |
| chr10_-_127195709 | 1.29 |
ENSMUST00000038217.7
ENSMUST00000130855.1 ENSMUST00000116229.1 ENSMUST00000144322.1 |
Dtx3
|
deltex 3 homolog (Drosophila) |
| chr9_-_18512885 | 1.26 |
ENSMUST00000034653.6
|
Muc16
|
mucin 16 |
| chr7_-_126799134 | 1.25 |
ENSMUST00000087566.4
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr2_+_91096744 | 1.23 |
ENSMUST00000132741.2
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr11_+_60469339 | 1.16 |
ENSMUST00000071880.2
ENSMUST00000081823.5 ENSMUST00000094135.2 |
Myo15
|
myosin XV |
| chr7_-_126799163 | 1.15 |
ENSMUST00000032934.5
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr4_-_58499398 | 1.11 |
ENSMUST00000107570.1
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr2_+_90677208 | 1.09 |
ENSMUST00000057481.6
|
Nup160
|
nucleoporin 160 |
| chr11_-_84819450 | 1.07 |
ENSMUST00000018549.7
|
Mrm1
|
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr19_-_24901309 | 1.07 |
ENSMUST00000058600.2
|
Foxd4
|
forkhead box D4 |
| chr3_+_105870898 | 1.05 |
ENSMUST00000010279.5
|
Adora3
|
adenosine A3 receptor |
| chr16_+_44765732 | 1.04 |
ENSMUST00000057488.8
|
Cd200r1
|
CD200 receptor 1 |
| chrX_+_35888808 | 1.04 |
ENSMUST00000033419.6
|
Dock11
|
dedicator of cytokinesis 11 |
| chr15_+_39198244 | 1.03 |
ENSMUST00000082054.5
ENSMUST00000042917.9 |
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr4_+_148804420 | 1.01 |
ENSMUST00000094464.3
ENSMUST00000122222.1 |
Casz1
|
castor zinc finger 1 |
| chr19_+_6401675 | 0.99 |
ENSMUST00000113471.1
ENSMUST00000113469.2 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr11_-_106160708 | 0.98 |
ENSMUST00000106875.1
|
Limd2
|
LIM domain containing 2 |
| chr5_-_137533297 | 0.96 |
ENSMUST00000111020.1
ENSMUST00000111023.1 |
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr7_+_90130227 | 0.96 |
ENSMUST00000049537.7
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr19_+_11404735 | 0.95 |
ENSMUST00000153546.1
|
Ms4a4c
|
membrane-spanning 4-domains, subfamily A, member 4C |
| chr5_-_134614953 | 0.92 |
ENSMUST00000036362.6
ENSMUST00000077636.4 |
Lat2
|
linker for activation of T cells family, member 2 |
| chr2_-_54085542 | 0.92 |
ENSMUST00000100089.2
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr3_+_87948666 | 0.91 |
ENSMUST00000005019.5
|
Crabp2
|
cellular retinoic acid binding protein II |
| chr11_-_103356324 | 0.90 |
ENSMUST00000136491.2
ENSMUST00000107023.2 |
Arhgap27
|
Rho GTPase activating protein 27 |
| chr11_+_69098937 | 0.89 |
ENSMUST00000021271.7
|
Per1
|
period circadian clock 1 |
| chr8_-_40634750 | 0.88 |
ENSMUST00000173957.1
|
Mtmr7
|
myotubularin related protein 7 |
| chr10_+_80016901 | 0.87 |
ENSMUST00000105373.1
|
Hmha1
|
histocompatibility (minor) HA-1 |
| chr7_-_92874196 | 0.87 |
ENSMUST00000032877.9
|
4632434I11Rik
|
RIKEN cDNA 4632434I11 gene |
| chr5_-_110839757 | 0.86 |
ENSMUST00000056937.5
|
Hscb
|
HscB iron-sulfur cluster co-chaperone homolog (E. coli) |
| chr2_+_24186469 | 0.86 |
ENSMUST00000057567.2
|
Il1f9
|
interleukin 1 family, member 9 |
| chr3_-_95307132 | 0.84 |
ENSMUST00000015846.2
|
Anxa9
|
annexin A9 |
| chr5_-_131538687 | 0.84 |
ENSMUST00000161374.1
|
Auts2
|
autism susceptibility candidate 2 |
| chr1_+_135836380 | 0.81 |
ENSMUST00000178204.1
|
Tnnt2
|
troponin T2, cardiac |
| chr8_-_27413937 | 0.80 |
ENSMUST00000033882.8
|
Chrna6
|
cholinergic receptor, nicotinic, alpha polypeptide 6 |
| chr4_-_139092958 | 0.80 |
ENSMUST00000042844.6
|
Nbl1
|
neuroblastoma, suppression of tumorigenicity 1 |
| chr5_-_110839575 | 0.79 |
ENSMUST00000145318.1
|
Hscb
|
HscB iron-sulfur cluster co-chaperone homolog (E. coli) |
| chr5_+_136083916 | 0.76 |
ENSMUST00000042135.7
|
Rasa4
|
RAS p21 protein activator 4 |
| chr19_+_55741810 | 0.76 |
ENSMUST00000111657.3
ENSMUST00000061496.9 ENSMUST00000041717.7 ENSMUST00000111662.4 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr5_+_120589003 | 0.75 |
ENSMUST00000069259.2
|
Iqcd
|
IQ motif containing D |
| chr9_-_32542861 | 0.75 |
ENSMUST00000183767.1
|
Fli1
|
Friend leukemia integration 1 |
| chr2_-_154569720 | 0.71 |
ENSMUST00000000894.5
|
E2f1
|
E2F transcription factor 1 |
| chr5_+_120589020 | 0.70 |
ENSMUST00000094391.4
|
Iqcd
|
IQ motif containing D |
| chr7_-_142061021 | 0.69 |
ENSMUST00000084418.2
|
Mob2
|
MOB kinase activator 2 |
| chr5_-_122989086 | 0.66 |
ENSMUST00000046073.9
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr5_-_137533212 | 0.65 |
ENSMUST00000143495.1
ENSMUST00000111038.1 |
Gnb2
Epo
|
guanine nucleotide binding protein (G protein), beta 2 erythropoietin |
| chr5_+_37338455 | 0.64 |
ENSMUST00000056365.8
|
Evc2
|
Ellis van Creveld syndrome 2 |
| chr11_+_70018421 | 0.63 |
ENSMUST00000108588.1
|
Dlg4
|
discs, large homolog 4 (Drosophila) |
| chr17_-_31512253 | 0.63 |
ENSMUST00000166626.1
|
Wdr4
|
WD repeat domain 4 |
| chr7_+_3289012 | 0.62 |
ENSMUST00000164553.1
|
Myadm
|
myeloid-associated differentiation marker |
| chr9_-_108083330 | 0.61 |
ENSMUST00000159372.1
ENSMUST00000160249.1 |
Rnf123
|
ring finger protein 123 |
| chr5_-_122989260 | 0.60 |
ENSMUST00000118027.1
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr14_+_61138445 | 0.59 |
ENSMUST00000089394.3
ENSMUST00000119509.1 |
Sacs
|
sacsin |
| chr7_+_101361250 | 0.58 |
ENSMUST00000137384.1
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr5_+_115327125 | 0.57 |
ENSMUST00000031513.7
|
Srsf9
|
serine/arginine-rich splicing factor 9 |
| chr10_-_25200110 | 0.57 |
ENSMUST00000100012.2
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr8_+_3631109 | 0.56 |
ENSMUST00000004745.8
|
Stxbp2
|
syntaxin binding protein 2 |
| chr17_+_17831004 | 0.55 |
ENSMUST00000172097.2
|
4930546H06Rik
|
RIKEN cDNA 4930546H06 gene |
| chr5_+_136084022 | 0.55 |
ENSMUST00000100570.3
|
Rasa4
|
RAS p21 protein activator 4 |
| chr4_+_109978004 | 0.54 |
ENSMUST00000061187.3
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2 |
| chrX_-_10216918 | 0.53 |
ENSMUST00000072393.2
ENSMUST00000044598.6 ENSMUST00000073392.4 ENSMUST00000115533.1 ENSMUST00000115532.1 |
Rpgr
|
retinitis pigmentosa GTPase regulator |
| chr12_+_53248677 | 0.53 |
ENSMUST00000101432.2
|
Npas3
|
neuronal PAS domain protein 3 |
| chr16_+_57353093 | 0.51 |
ENSMUST00000159816.1
|
Filip1l
|
filamin A interacting protein 1-like |
| chr16_+_30008657 | 0.51 |
ENSMUST00000181485.1
|
4632428C04Rik
|
RIKEN cDNA 4632428C04 gene |
| chr11_-_59029509 | 0.51 |
ENSMUST00000127937.1
|
Obscn
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chrX_+_166344692 | 0.47 |
ENSMUST00000112223.1
ENSMUST00000112224.1 ENSMUST00000112229.2 ENSMUST00000112228.1 ENSMUST00000112227.2 ENSMUST00000112226.2 |
Gpm6b
|
glycoprotein m6b |
| chr7_-_29518566 | 0.46 |
ENSMUST00000181975.1
|
Sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chrX_-_10216437 | 0.45 |
ENSMUST00000115534.1
|
Rpgr
|
retinitis pigmentosa GTPase regulator |
| chr7_-_45016224 | 0.45 |
ENSMUST00000085383.2
|
Scaf1
|
SR-related CTD-associated factor 1 |
| chr1_-_93801840 | 0.45 |
ENSMUST00000112890.2
ENSMUST00000027503.7 |
Dtymk
|
deoxythymidylate kinase |
| chr2_-_164071124 | 0.44 |
ENSMUST00000109384.3
|
Tomm34
|
translocase of outer mitochondrial membrane 34 |
| chr13_+_22327911 | 0.43 |
ENSMUST00000091734.1
|
Vmn1r197
|
vomeronasal 1 receptor 197 |
| chr15_-_98778150 | 0.43 |
ENSMUST00000023732.5
|
Wnt10b
|
wingless related MMTV integration site 10b |
| chr5_-_137533170 | 0.42 |
ENSMUST00000168746.1
ENSMUST00000170293.1 |
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr3_+_87971071 | 0.39 |
ENSMUST00000090973.5
|
Nes
|
nestin |
| chr2_-_57124003 | 0.38 |
ENSMUST00000112629.1
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr3_-_66981279 | 0.38 |
ENSMUST00000162098.2
|
Shox2
|
short stature homeobox 2 |
| chr15_-_85581809 | 0.38 |
ENSMUST00000023015.7
|
Wnt7b
|
wingless-related MMTV integration site 7B |
| chr7_-_19715395 | 0.37 |
ENSMUST00000032555.9
ENSMUST00000093552.5 |
Tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr10_+_36974536 | 0.37 |
ENSMUST00000019911.7
|
Hdac2
|
histone deacetylase 2 |
| chr13_+_23738804 | 0.37 |
ENSMUST00000040914.1
|
Hist1h1c
|
histone cluster 1, H1c |
| chr3_+_87971129 | 0.36 |
ENSMUST00000160694.1
|
Nes
|
nestin |
| chr2_-_26294550 | 0.35 |
ENSMUST00000057224.3
|
4932418E24Rik
|
RIKEN cDNA 4932418E24 gene |
| chr10_+_36974558 | 0.35 |
ENSMUST00000105510.1
|
Hdac2
|
histone deacetylase 2 |
| chr5_+_140419248 | 0.34 |
ENSMUST00000100507.3
|
Eif3b
|
eukaryotic translation initiation factor 3, subunit B |
| chr11_+_77493408 | 0.33 |
ENSMUST00000037285.3
ENSMUST00000100812.3 |
Git1
|
G protein-coupled receptor kinase-interactor 1 |
| chr16_+_17561885 | 0.32 |
ENSMUST00000171002.1
ENSMUST00000023441.4 |
P2rx6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr1_+_170232749 | 0.32 |
ENSMUST00000162752.1
|
Sh2d1b2
|
SH2 domain protein 1B2 |
| chr7_+_25282784 | 0.31 |
ENSMUST00000165239.1
|
Cic
|
capicua homolog (Drosophila) |
| chr2_+_164656043 | 0.31 |
ENSMUST00000094344.5
|
Wfdc10
|
WAP four-disulfide core domain 10 |
| chr19_+_29410919 | 0.31 |
ENSMUST00000112576.2
|
Pdcd1lg2
|
programmed cell death 1 ligand 2 |
| chr2_+_172979827 | 0.29 |
ENSMUST00000109125.1
ENSMUST00000050442.9 ENSMUST00000109126.3 |
Spo11
|
SPO11 meiotic protein covalently bound to DSB homolog (S. cerevisiae) |
| chr4_-_117914968 | 0.29 |
ENSMUST00000036156.5
|
Ipo13
|
importin 13 |
| chr5_+_30105161 | 0.28 |
ENSMUST00000058045.4
|
Gareml
|
GRB2 associated, regulator of MAPK1-like |
| chr17_+_29679247 | 0.28 |
ENSMUST00000129864.1
|
Cmtr1
|
cap methyltransferase 1 |
| chr1_-_152766281 | 0.28 |
ENSMUST00000111859.1
ENSMUST00000148865.1 |
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr12_-_80643799 | 0.28 |
ENSMUST00000166931.1
|
Erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr14_-_76520850 | 0.26 |
ENSMUST00000181972.1
|
4930444M15Rik
|
RIKEN cDNA 4930444M15 gene |
| chr2_-_3512746 | 0.25 |
ENSMUST00000056700.7
ENSMUST00000027961.5 |
Hspa14
Hspa14
|
heat shock protein 14 heat shock protein 14 |
| chr13_+_109632760 | 0.25 |
ENSMUST00000135275.1
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr9_-_106465940 | 0.25 |
ENSMUST00000164834.1
|
Gpr62
|
G protein-coupled receptor 62 |
| chr5_+_77004055 | 0.24 |
ENSMUST00000071199.2
|
Arl9
|
ADP-ribosylation factor-like 9 |
| chr2_+_70039114 | 0.23 |
ENSMUST00000060208.4
|
Myo3b
|
myosin IIIB |
| chr7_-_35647441 | 0.23 |
ENSMUST00000118501.1
|
Pdcd5
|
programmed cell death 5 |
| chr1_-_192092540 | 0.23 |
ENSMUST00000085573.6
|
Traf5
|
TNF receptor-associated factor 5 |
| chr9_-_20336094 | 0.23 |
ENSMUST00000086473.3
|
Olfr18
|
olfactory receptor 18 |
| chr8_-_120101473 | 0.21 |
ENSMUST00000034280.7
|
Zdhhc7
|
zinc finger, DHHC domain containing 7 |
| chr6_-_116716888 | 0.21 |
ENSMUST00000056623.6
|
Tmem72
|
transmembrane protein 72 |
| chr1_+_39900883 | 0.20 |
ENSMUST00000163854.2
ENSMUST00000168431.1 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr14_+_34086008 | 0.20 |
ENSMUST00000120077.1
|
Anxa8
|
annexin A8 |
| chrX_-_148615153 | 0.19 |
ENSMUST00000112755.2
|
Gm8334
|
predicted gene 8334 |
| chr16_+_57353271 | 0.18 |
ENSMUST00000099667.2
|
Filip1l
|
filamin A interacting protein 1-like |
| chr4_-_62360436 | 0.17 |
ENSMUST00000084527.3
ENSMUST00000098033.3 |
Fkbp15
|
FK506 binding protein 15 |
| chr7_+_5020561 | 0.17 |
ENSMUST00000085427.3
|
Zfp865
|
zinc finger protein 865 |
| chr14_+_60378242 | 0.17 |
ENSMUST00000022561.6
|
Amer2
|
APC membrane recruitment 2 |
| chr4_+_152158867 | 0.16 |
ENSMUST00000030782.1
|
Hes2
|
hairy and enhancer of split 2 (Drosophila) |
| chr16_-_32099697 | 0.16 |
ENSMUST00000155966.1
ENSMUST00000096109.4 |
Pigx
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr7_-_114636299 | 0.16 |
ENSMUST00000032906.4
ENSMUST00000032907.7 |
Calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr6_+_120666388 | 0.15 |
ENSMUST00000112686.1
|
Cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr3_-_90509450 | 0.15 |
ENSMUST00000107343.1
ENSMUST00000001043.7 ENSMUST00000107344.1 ENSMUST00000076639.4 ENSMUST00000107346.1 ENSMUST00000146740.1 ENSMUST00000107342.1 ENSMUST00000049937.6 |
Chtop
|
chromatin target of PRMT1 |
| chr7_-_30193098 | 0.14 |
ENSMUST00000108196.1
|
Capns1
|
calpain, small subunit 1 |
| chr7_+_5020376 | 0.14 |
ENSMUST00000076251.4
|
Zfp865
|
zinc finger protein 865 |
| chr4_+_41569775 | 0.13 |
ENSMUST00000102963.3
|
Dnaic1
|
dynein, axonemal, intermediate chain 1 |
| chr6_-_4747019 | 0.13 |
ENSMUST00000126151.1
ENSMUST00000133306.1 ENSMUST00000123907.1 |
Sgce
|
sarcoglycan, epsilon |
| chr9_-_54647199 | 0.13 |
ENSMUST00000128163.1
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr7_+_28881656 | 0.12 |
ENSMUST00000066880.4
|
Capn12
|
calpain 12 |
| chr1_-_153487639 | 0.12 |
ENSMUST00000042141.5
|
Dhx9
|
DEAH (Asp-Glu-Ala-His) box polypeptide 9 |
| chr2_+_4976113 | 0.12 |
ENSMUST00000167607.1
ENSMUST00000115010.2 |
Ucma
|
upper zone of growth plate and cartilage matrix associated |
| chr10_+_127078886 | 0.11 |
ENSMUST00000039259.6
|
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr12_-_11150305 | 0.10 |
ENSMUST00000055673.1
|
Kcns3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr12_+_73123709 | 0.10 |
ENSMUST00000021523.6
|
Mnat1
|
menage a trois 1 |
| chr17_+_35016576 | 0.09 |
ENSMUST00000007245.1
ENSMUST00000172499.1 |
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr5_-_123524124 | 0.09 |
ENSMUST00000111586.1
ENSMUST00000031385.6 ENSMUST00000111587.3 |
Diablo
|
diablo homolog (Drosophila) |
| chr2_+_83644435 | 0.09 |
ENSMUST00000081591.6
|
Zc3h15
|
zinc finger CCCH-type containing 15 |
| chr8_-_24596960 | 0.08 |
ENSMUST00000033956.6
|
Ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr2_+_174330006 | 0.08 |
ENSMUST00000109085.1
ENSMUST00000109087.1 ENSMUST00000109084.1 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr1_-_74284636 | 0.07 |
ENSMUST00000178235.1
ENSMUST00000006462.7 |
Aamp
|
angio-associated migratory protein |
| chr6_-_51469869 | 0.06 |
ENSMUST00000114459.1
ENSMUST00000069949.6 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr9_+_89909775 | 0.06 |
ENSMUST00000034912.4
ENSMUST00000034909.4 |
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr7_+_5057161 | 0.06 |
ENSMUST00000045543.5
|
Ccdc106
|
coiled-coil domain containing 106 |
| chr7_-_35647127 | 0.05 |
ENSMUST00000120714.1
|
Pdcd5
|
programmed cell death 5 |
| chr16_+_16983382 | 0.05 |
ENSMUST00000069107.7
ENSMUST00000023462.6 |
Mapk1
|
mitogen-activated protein kinase 1 |
| chr2_-_57113053 | 0.04 |
ENSMUST00000112627.1
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr7_+_30459713 | 0.02 |
ENSMUST00000006825.8
|
Nphs1
|
nephrosis 1, nephrin |
| chr5_+_115279666 | 0.01 |
ENSMUST00000040421.4
|
Coq5
|
coenzyme Q5 homolog, methyltransferase (yeast) |
| chr2_+_4976176 | 0.01 |
ENSMUST00000027978.1
|
Ucma
|
upper zone of growth plate and cartilage matrix associated |
| chr11_+_46436925 | 0.01 |
ENSMUST00000152119.1
ENSMUST00000140027.1 ENSMUST00000020665.6 ENSMUST00000170928.1 ENSMUST00000109231.1 ENSMUST00000109232.3 ENSMUST00000128940.1 |
Med7
|
mediator complex subunit 7 |
| chr5_+_94873797 | 0.01 |
ENSMUST00000180076.1
|
Gm3183
|
predicted gene 3183 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Glis2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.2 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 1.3 | 3.9 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 1.2 | 3.6 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) regulation of cGMP-mediated signaling(GO:0010752) |
| 1.2 | 3.5 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.8 | 4.8 | GO:0032796 | uropod organization(GO:0032796) |
| 0.8 | 3.2 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.8 | 6.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.4 | 1.3 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.4 | 6.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.4 | 1.7 | GO:0060032 | notochord regression(GO:0060032) |
| 0.3 | 1.0 | GO:1902870 | camera-type eye photoreceptor cell fate commitment(GO:0060220) negative regulation of neural retina development(GO:0061076) negative regulation of retina development in camera-type eye(GO:1902867) negative regulation of amacrine cell differentiation(GO:1902870) |
| 0.3 | 0.8 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.3 | 1.1 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.3 | 0.8 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.3 | 7.6 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.3 | 2.5 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.3 | 1.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.2 | 1.2 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.2 | 1.7 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.2 | 1.9 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 1.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 3.7 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 0.4 | GO:0046075 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.2 | 0.9 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.2 | 0.4 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.2 | 1.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 | 1.3 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.2 | 0.8 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.2 | 1.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 1.0 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.2 | 3.7 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.2 | 0.7 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.2 | 0.9 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.2 | 2.6 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.2 | 1.9 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.2 | 2.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 0.6 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.7 | GO:0035983 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.1 | 0.4 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 2.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.4 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.1 | 0.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 1.0 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 2.4 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 0.3 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 0.3 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.3 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.1 | 0.6 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.6 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 0.8 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.1 | 0.5 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 1.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.3 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.1 | 0.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.8 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.1 | 0.6 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 0.2 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.9 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 1.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 1.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.6 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.9 | GO:0043303 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 0.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.6 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.3 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.0 | 1.1 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 1.0 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.5 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.5 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.3 | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.0 | 0.3 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.8 | 6.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.7 | 10.7 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.4 | 3.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 1.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 1.0 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.3 | 3.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 2.6 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.2 | 5.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.7 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 0.8 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 2.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.6 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 1.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 1.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.5 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.8 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 1.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 1.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 4.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 2.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 1.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.0 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 1.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 3.5 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 2.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.1 | GO:0043198 | dendritic shaft(GO:0043198) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.9 | 3.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.9 | 3.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.6 | 1.9 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.4 | 6.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 3.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.3 | 1.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.3 | 2.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 2.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.3 | 0.8 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 2.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 4.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 0.6 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.2 | 0.6 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.2 | 1.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.2 | 0.7 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) Krueppel-associated box domain binding(GO:0035851) |
| 0.2 | 10.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.2 | 1.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.4 | GO:0009041 | uridylate kinase activity(GO:0009041) nucleoside phosphate kinase activity(GO:0050145) |
| 0.1 | 0.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.8 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 1.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 3.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.3 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 2.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.4 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.1 | 4.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.6 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.8 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.6 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 0.3 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 1.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 1.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.9 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.3 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 1.3 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 0.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 1.9 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 2.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 0.0 | 0.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 1.1 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.5 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.6 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 1.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 3.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 3.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 10.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 3.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 3.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 3.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 2.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 2.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 2.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 2.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 3.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 2.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 3.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 3.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 5.6 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 1.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.9 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 3.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.7 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 0.8 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 2.4 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 2.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 3.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 1.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 1.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |