Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
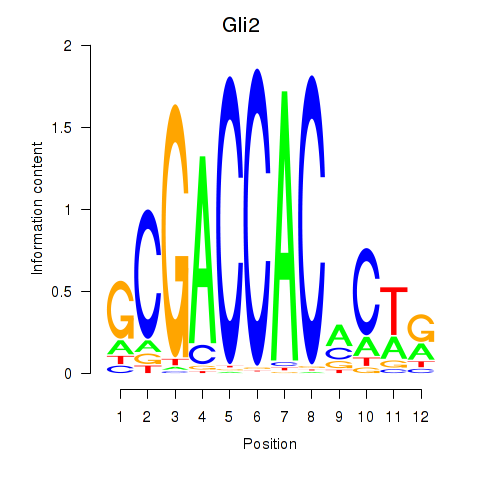
Results for Gli2
Z-value: 1.48
Transcription factors associated with Gli2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gli2
|
ENSMUSG00000048402.8 | GLI-Kruppel family member GLI2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gli2 | mm10_v2_chr1_-_119053619_119053638 | -0.82 | 6.3e-10 | Click! |
Activity profile of Gli2 motif
Sorted Z-values of Gli2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_53213447 | 4.94 |
ENSMUST00000031090.6
|
Sel1l3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr1_-_51915901 | 4.65 |
ENSMUST00000018561.7
ENSMUST00000114537.2 |
Myo1b
|
myosin IB |
| chr1_-_51915968 | 4.11 |
ENSMUST00000046390.7
|
Myo1b
|
myosin IB |
| chr4_-_154899077 | 4.10 |
ENSMUST00000030935.3
ENSMUST00000132281.1 |
Fam213b
|
family with sequence similarity 213, member B |
| chr3_+_97628804 | 4.02 |
ENSMUST00000107050.1
ENSMUST00000029729.8 ENSMUST00000107049.1 |
Fmo5
|
flavin containing monooxygenase 5 |
| chr1_-_180199587 | 3.82 |
ENSMUST00000161743.1
|
Adck3
|
aarF domain containing kinase 3 |
| chr9_+_107576915 | 3.56 |
ENSMUST00000112387.2
ENSMUST00000123005.1 ENSMUST00000010195.7 ENSMUST00000144392.1 |
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr17_-_24209377 | 3.43 |
ENSMUST00000024931.4
|
Ntn3
|
netrin 3 |
| chr7_-_81454751 | 3.41 |
ENSMUST00000098331.3
ENSMUST00000178892.1 |
Cpeb1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr8_+_76902277 | 2.89 |
ENSMUST00000109912.1
ENSMUST00000128862.1 ENSMUST00000109911.1 |
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr10_-_114801364 | 2.58 |
ENSMUST00000061632.7
|
Trhde
|
TRH-degrading enzyme |
| chr4_-_121098190 | 2.56 |
ENSMUST00000058754.2
|
Zmpste24
|
zinc metallopeptidase, STE24 |
| chr12_-_70227622 | 2.56 |
ENSMUST00000071250.6
|
Pygl
|
liver glycogen phosphorylase |
| chr9_+_100643755 | 2.53 |
ENSMUST00000133388.1
|
Stag1
|
stromal antigen 1 |
| chrX_+_161717498 | 2.52 |
ENSMUST00000061514.7
|
Rai2
|
retinoic acid induced 2 |
| chr2_+_69897255 | 2.40 |
ENSMUST00000131553.1
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr5_-_139819906 | 2.34 |
ENSMUST00000147328.1
|
Tmem184a
|
transmembrane protein 184a |
| chr9_+_100643605 | 2.19 |
ENSMUST00000041418.6
|
Stag1
|
stromal antigen 1 |
| chr8_+_127064107 | 2.14 |
ENSMUST00000162536.1
ENSMUST00000026921.6 ENSMUST00000162665.1 ENSMUST00000160766.1 ENSMUST00000162602.1 ENSMUST00000162531.1 ENSMUST00000160581.1 ENSMUST00000161355.1 ENSMUST00000159537.1 |
Pard3
|
par-3 (partitioning defective 3) homolog (C. elegans) |
| chr13_-_54611274 | 2.08 |
ENSMUST00000049575.7
|
Cltb
|
clathrin, light polypeptide (Lcb) |
| chr8_+_127064022 | 2.01 |
ENSMUST00000160272.1
ENSMUST00000079777.5 ENSMUST00000162907.1 |
Pard3
|
par-3 (partitioning defective 3) homolog (C. elegans) |
| chr19_-_43674844 | 1.94 |
ENSMUST00000046038.7
|
Slc25a28
|
solute carrier family 25, member 28 |
| chr8_+_127063893 | 1.94 |
ENSMUST00000162309.1
|
Pard3
|
par-3 (partitioning defective 3) homolog (C. elegans) |
| chr4_-_46536096 | 1.92 |
ENSMUST00000102924.2
|
Trim14
|
tripartite motif-containing 14 |
| chr9_-_43239816 | 1.92 |
ENSMUST00000034512.5
|
Oaf
|
OAF homolog (Drosophila) |
| chr4_-_46536134 | 1.87 |
ENSMUST00000046897.6
|
Trim14
|
tripartite motif-containing 14 |
| chrX_-_9662950 | 1.86 |
ENSMUST00000033519.2
|
Dynlt3
|
dynein light chain Tctex-type 3 |
| chr6_+_113471481 | 1.83 |
ENSMUST00000113062.1
|
Il17rc
|
interleukin 17 receptor C |
| chr10_+_80151154 | 1.82 |
ENSMUST00000146516.1
ENSMUST00000144526.1 |
Midn
|
midnolin |
| chr16_-_17132377 | 1.79 |
ENSMUST00000023453.7
|
Sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr13_-_54611332 | 1.76 |
ENSMUST00000091609.4
|
Cltb
|
clathrin, light polypeptide (Lcb) |
| chr11_-_121388186 | 1.70 |
ENSMUST00000106107.2
|
Rab40b
|
Rab40b, member RAS oncogene family |
| chr1_-_90967667 | 1.70 |
ENSMUST00000131428.1
|
Rab17
|
RAB17, member RAS oncogene family |
| chr2_-_157566319 | 1.70 |
ENSMUST00000109528.2
ENSMUST00000088494.2 |
Blcap
|
bladder cancer associated protein homolog (human) |
| chr6_+_113471427 | 1.69 |
ENSMUST00000058300.7
|
Il17rc
|
interleukin 17 receptor C |
| chr9_+_100643448 | 1.64 |
ENSMUST00000146312.1
ENSMUST00000129269.1 |
Stag1
|
stromal antigen 1 |
| chr2_-_121380940 | 1.64 |
ENSMUST00000038389.8
|
Strc
|
stereocilin |
| chr5_-_135962275 | 1.61 |
ENSMUST00000054895.3
|
Srcrb4d
|
scavenger receptor cysteine rich domain containing, group B (4 domains) |
| chr9_+_109095427 | 1.54 |
ENSMUST00000072093.6
|
Plxnb1
|
plexin B1 |
| chr7_+_35119285 | 1.53 |
ENSMUST00000042985.9
|
Cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr7_-_144738520 | 1.44 |
ENSMUST00000118556.2
ENSMUST00000033393.8 |
Ano1
|
anoctamin 1, calcium activated chloride channel |
| chr19_+_6047081 | 1.41 |
ENSMUST00000025723.8
|
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr2_+_140395309 | 1.38 |
ENSMUST00000110067.1
ENSMUST00000110064.1 ENSMUST00000110063.1 ENSMUST00000110062.1 ENSMUST00000078027.5 ENSMUST00000043836.7 |
Macrod2
|
MACRO domain containing 2 |
| chr7_-_144738478 | 1.35 |
ENSMUST00000121758.1
|
Ano1
|
anoctamin 1, calcium activated chloride channel |
| chr17_+_28691342 | 1.30 |
ENSMUST00000114758.1
ENSMUST00000004990.6 ENSMUST00000062694.8 ENSMUST00000114754.1 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr2_-_37703275 | 1.29 |
ENSMUST00000072186.5
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr19_+_6047055 | 1.29 |
ENSMUST00000134667.1
|
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr7_+_81571246 | 1.28 |
ENSMUST00000165460.1
|
Whamm
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr6_+_43265582 | 1.28 |
ENSMUST00000031750.7
|
Arhgef5
|
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr12_-_31950170 | 1.27 |
ENSMUST00000176520.1
|
Hbp1
|
high mobility group box transcription factor 1 |
| chrX_-_74023908 | 1.26 |
ENSMUST00000033769.8
ENSMUST00000114352.1 ENSMUST00000068286.5 ENSMUST00000114360.3 ENSMUST00000114354.3 |
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr10_+_83543941 | 1.24 |
ENSMUST00000038388.5
|
A230046K03Rik
|
RIKEN cDNA A230046K03 gene |
| chr1_-_175625580 | 1.24 |
ENSMUST00000027810.7
|
Fh1
|
fumarate hydratase 1 |
| chr11_+_29526423 | 1.18 |
ENSMUST00000136351.1
ENSMUST00000020749.6 ENSMUST00000144321.1 ENSMUST00000093239.4 |
Mtif2
|
mitochondrial translational initiation factor 2 |
| chrX_-_141725181 | 1.17 |
ENSMUST00000067841.7
|
Irs4
|
insulin receptor substrate 4 |
| chr11_+_29526407 | 1.17 |
ENSMUST00000133452.1
|
Mtif2
|
mitochondrial translational initiation factor 2 |
| chr7_-_121035096 | 1.14 |
ENSMUST00000065740.2
|
Gm9905
|
predicted gene 9905 |
| chr8_-_13890233 | 1.13 |
ENSMUST00000033839.7
|
Coprs
|
coordinator of PRMT5, differentiation stimulator |
| chr15_+_41447438 | 1.13 |
ENSMUST00000110297.2
ENSMUST00000090096.4 |
Oxr1
|
oxidation resistance 1 |
| chr7_+_121707189 | 1.12 |
ENSMUST00000065310.2
|
1700069B07Rik
|
RIKEN cDNA 1700069B07 gene |
| chr7_+_102578469 | 1.09 |
ENSMUST00000098225.2
|
Olfr550
|
olfactory receptor 550 |
| chr17_+_28691419 | 1.09 |
ENSMUST00000124886.1
|
Mapk14
|
mitogen-activated protein kinase 14 |
| chr4_-_140648736 | 1.09 |
ENSMUST00000039204.3
ENSMUST00000105799.1 ENSMUST00000097820.2 |
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr5_-_135962265 | 1.09 |
ENSMUST00000111150.1
|
Srcrb4d
|
scavenger receptor cysteine rich domain containing, group B (4 domains) |
| chr2_+_49451486 | 1.08 |
ENSMUST00000092123.4
|
Epc2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr7_-_133123312 | 1.07 |
ENSMUST00000171022.1
ENSMUST00000163601.1 ENSMUST00000165534.1 ENSMUST00000033269.8 ENSMUST00000124096.1 |
Ctbp2
Fgfr2
|
C-terminal binding protein 2 fibroblast growth factor receptor 2 |
| chr9_+_62342449 | 1.07 |
ENSMUST00000156461.1
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr4_+_43401232 | 1.06 |
ENSMUST00000125399.1
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr4_-_19708922 | 1.06 |
ENSMUST00000108246.2
|
Wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chrX_+_161717055 | 1.05 |
ENSMUST00000112338.1
|
Rai2
|
retinoic acid induced 2 |
| chr2_+_130576170 | 1.02 |
ENSMUST00000028764.5
|
Oxt
|
oxytocin |
| chr1_+_167349976 | 1.01 |
ENSMUST00000028004.9
|
Aldh9a1
|
aldehyde dehydrogenase 9, subfamily A1 |
| chr12_-_31950210 | 0.99 |
ENSMUST00000176084.1
ENSMUST00000176103.1 ENSMUST00000167458.2 |
Hbp1
|
high mobility group box transcription factor 1 |
| chr2_+_160731684 | 0.99 |
ENSMUST00000174885.1
ENSMUST00000109462.1 |
Plcg1
|
phospholipase C, gamma 1 |
| chr6_-_124464772 | 0.98 |
ENSMUST00000008297.4
|
Clstn3
|
calsyntenin 3 |
| chr11_+_70844745 | 0.97 |
ENSMUST00000076270.6
ENSMUST00000179114.1 ENSMUST00000100928.4 ENSMUST00000177731.1 ENSMUST00000108533.3 ENSMUST00000081362.6 ENSMUST00000178245.1 |
Rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr18_-_35215008 | 0.96 |
ENSMUST00000091636.3
|
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr17_+_24669730 | 0.95 |
ENSMUST00000047179.5
|
Zfp598
|
zinc finger protein 598 |
| chr16_-_31314804 | 0.93 |
ENSMUST00000115230.1
ENSMUST00000130560.1 |
Apod
|
apolipoprotein D |
| chr11_+_72796254 | 0.90 |
ENSMUST00000069395.5
|
Zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr10_-_34127955 | 0.90 |
ENSMUST00000062784.6
|
Fam26f
|
family with sequence similarity 26, member F |
| chr8_-_67515606 | 0.90 |
ENSMUST00000032981.5
|
Gm9755
|
predicted pseudogene 9755 |
| chr5_-_71095765 | 0.88 |
ENSMUST00000000572.5
|
Gabra2
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 2 |
| chr11_+_72796164 | 0.87 |
ENSMUST00000172220.1
|
Zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr11_+_73329727 | 0.85 |
ENSMUST00000092926.4
ENSMUST00000117445.1 |
Spata22
|
spermatogenesis associated 22 |
| chr7_-_116308241 | 0.85 |
ENSMUST00000183057.1
ENSMUST00000182487.1 ENSMUST00000181998.1 |
Plekha7
|
pleckstrin homology domain containing, family A member 7 |
| chr8_+_85060055 | 0.84 |
ENSMUST00000095220.3
|
Fbxw9
|
F-box and WD-40 domain protein 9 |
| chr5_-_134456227 | 0.82 |
ENSMUST00000111244.1
|
Gtf2ird1
|
general transcription factor II I repeat domain-containing 1 |
| chr2_+_164486455 | 0.82 |
ENSMUST00000069385.8
ENSMUST00000143690.1 |
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr7_-_137314394 | 0.81 |
ENSMUST00000168203.1
ENSMUST00000106118.2 ENSMUST00000169486.2 ENSMUST00000033378.5 |
Ebf3
|
early B cell factor 3 |
| chr9_+_45055211 | 0.81 |
ENSMUST00000114663.2
|
Mpzl3
|
myelin protein zero-like 3 |
| chr9_+_55149364 | 0.80 |
ENSMUST00000121677.1
|
Ube2q2
|
ubiquitin-conjugating enzyme E2Q (putative) 2 |
| chrX_-_95956890 | 0.80 |
ENSMUST00000079987.6
ENSMUST00000113864.2 |
Las1l
|
LAS1-like (S. cerevisiae) |
| chr11_+_95712673 | 0.75 |
ENSMUST00000107717.1
|
Zfp652
|
zinc finger protein 652 |
| chr18_+_61555689 | 0.75 |
ENSMUST00000167187.1
|
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr17_-_34214459 | 0.75 |
ENSMUST00000121995.1
|
Gm15821
|
predicted gene 15821 |
| chr18_+_38250240 | 0.75 |
ENSMUST00000025314.5
|
0610009O20Rik
|
RIKEN cDNA 0610009O20 gene |
| chr4_-_126256226 | 0.74 |
ENSMUST00000122129.1
ENSMUST00000061143.8 ENSMUST00000106132.2 |
Map7d1
|
MAP7 domain containing 1 |
| chr3_+_94837533 | 0.73 |
ENSMUST00000107270.2
|
Pogz
|
pogo transposable element with ZNF domain |
| chr2_-_120609500 | 0.72 |
ENSMUST00000133612.1
ENSMUST00000102498.1 ENSMUST00000102499.1 |
Lrrc57
|
leucine rich repeat containing 57 |
| chr10_+_76531593 | 0.71 |
ENSMUST00000048678.6
|
Lss
|
lanosterol synthase |
| chr17_+_24632671 | 0.71 |
ENSMUST00000047611.2
|
Nthl1
|
nth (endonuclease III)-like 1 (E.coli) |
| chr3_+_127633134 | 0.71 |
ENSMUST00000029587.7
|
Neurog2
|
neurogenin 2 |
| chr5_-_24527276 | 0.71 |
ENSMUST00000088311.4
|
Gbx1
|
gastrulation brain homeobox 1 |
| chr2_-_120609319 | 0.66 |
ENSMUST00000102497.3
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr8_-_27174623 | 0.64 |
ENSMUST00000033878.6
ENSMUST00000054212.6 |
Rab11fip1
|
RAB11 family interacting protein 1 (class I) |
| chr15_-_78718113 | 0.61 |
ENSMUST00000088592.4
|
Elfn2
|
leucine rich repeat and fibronectin type III, extracellular 2 |
| chr14_+_25607797 | 0.61 |
ENSMUST00000160229.1
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr8_+_70754679 | 0.60 |
ENSMUST00000110093.2
ENSMUST00000143118.1 ENSMUST00000034301.5 ENSMUST00000110090.1 |
Rab3a
|
RAB3A, member RAS oncogene family |
| chr17_-_25497288 | 0.59 |
ENSMUST00000165183.2
ENSMUST00000051864.4 |
Sstr5
|
somatostatin receptor 5 |
| chr16_-_4077778 | 0.59 |
ENSMUST00000006137.8
|
Trap1
|
TNF receptor-associated protein 1 |
| chr14_-_64949838 | 0.58 |
ENSMUST00000067843.3
ENSMUST00000176489.1 ENSMUST00000175905.1 ENSMUST00000022544.7 ENSMUST00000175744.1 ENSMUST00000176128.1 |
Hmbox1
|
homeobox containing 1 |
| chr11_-_94704499 | 0.58 |
ENSMUST00000069852.1
|
Gm11541
|
predicted gene 11541 |
| chr10_-_17947997 | 0.57 |
ENSMUST00000037879.6
|
Heca
|
headcase homolog (Drosophila) |
| chr19_-_8880883 | 0.57 |
ENSMUST00000096253.5
|
AI462493
|
expressed sequence AI462493 |
| chr11_+_116030304 | 0.55 |
ENSMUST00000021116.5
ENSMUST00000106452.1 |
Unk
|
unkempt homolog (Drosophila) |
| chr10_-_127263346 | 0.55 |
ENSMUST00000099172.3
|
Kif5a
|
kinesin family member 5A |
| chr9_+_45055166 | 0.54 |
ENSMUST00000114664.1
ENSMUST00000093856.3 |
Mpzl3
|
myelin protein zero-like 3 |
| chr4_-_135971894 | 0.53 |
ENSMUST00000105852.1
|
Lypla2
|
lysophospholipase 2 |
| chr2_+_121956411 | 0.53 |
ENSMUST00000110578.1
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr17_-_56609689 | 0.53 |
ENSMUST00000052832.5
|
2410015M20Rik
|
RIKEN cDNA 2410015M20 gene |
| chr2_-_120609283 | 0.52 |
ENSMUST00000102496.1
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr8_+_107436355 | 0.52 |
ENSMUST00000166615.1
|
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chrX_-_167382747 | 0.51 |
ENSMUST00000026839.4
|
Prps2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr9_+_107928440 | 0.51 |
ENSMUST00000085073.1
|
Actl11
|
actin-like 11 |
| chr11_-_68871848 | 0.50 |
ENSMUST00000101017.2
|
Ndel1
|
nuclear distribution gene E-like homolog 1 (A. nidulans) |
| chr11_-_107794557 | 0.50 |
ENSMUST00000021066.3
|
Cacng4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr11_+_117308135 | 0.50 |
ENSMUST00000153668.1
|
Sept9
|
septin 9 |
| chr7_-_65371210 | 0.49 |
ENSMUST00000102592.3
|
Tjp1
|
tight junction protein 1 |
| chr11_+_115912001 | 0.49 |
ENSMUST00000132961.1
|
Smim6
|
small integral membrane protein 6 |
| chr2_+_120609383 | 0.47 |
ENSMUST00000124187.1
|
Haus2
|
HAUS augmin-like complex, subunit 2 |
| chr11_-_42000532 | 0.47 |
ENSMUST00000070735.3
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr12_+_72085847 | 0.41 |
ENSMUST00000117449.1
ENSMUST00000057257.8 |
Jkamp
|
JNK1/MAPK8-associated membrane protein |
| chr3_+_95427575 | 0.41 |
ENSMUST00000181809.1
|
Gm4349
|
predicted gene 4349 |
| chr4_+_155831630 | 0.41 |
ENSMUST00000105592.1
ENSMUST00000105591.1 |
Aurkaip1
|
aurora kinase A interacting protein 1 |
| chr8_+_119612747 | 0.40 |
ENSMUST00000098361.2
|
Adad2
|
adenosine deaminase domain containing 2 |
| chr11_-_42000284 | 0.40 |
ENSMUST00000109292.2
ENSMUST00000109290.1 |
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr11_+_70562898 | 0.39 |
ENSMUST00000102559.4
|
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr12_-_31950535 | 0.39 |
ENSMUST00000172314.2
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr16_-_88056176 | 0.36 |
ENSMUST00000072256.5
ENSMUST00000023652.8 ENSMUST00000114137.1 |
Grik1
|
glutamate receptor, ionotropic, kainate 1 |
| chr3_-_135608221 | 0.36 |
ENSMUST00000132668.1
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
| chr11_+_75532127 | 0.35 |
ENSMUST00000127226.1
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr9_-_69760924 | 0.34 |
ENSMUST00000071281.4
|
Foxb1
|
forkhead box B1 |
| chrX_+_99003224 | 0.32 |
ENSMUST00000149999.1
|
Stard8
|
START domain containing 8 |
| chr18_+_61555258 | 0.32 |
ENSMUST00000165123.1
|
Csnk1a1
|
casein kinase 1, alpha 1 |
| chrX_+_137049586 | 0.30 |
ENSMUST00000047852.7
|
Fam199x
|
family with sequence similarity 199, X-linked |
| chr1_+_71652837 | 0.30 |
ENSMUST00000097699.2
|
Apol7d
|
apolipoprotein L 7d |
| chr7_+_141328122 | 0.30 |
ENSMUST00000133012.1
ENSMUST00000026578.7 |
Tmem80
|
transmembrane protein 80 |
| chr18_+_61555308 | 0.29 |
ENSMUST00000165721.1
ENSMUST00000115246.2 ENSMUST00000166990.1 ENSMUST00000163205.1 ENSMUST00000170862.1 |
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr6_+_120666388 | 0.28 |
ENSMUST00000112686.1
|
Cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr5_-_20882072 | 0.27 |
ENSMUST00000118174.1
|
Phtf2
|
putative homeodomain transcription factor 2 |
| chr3_+_9250602 | 0.26 |
ENSMUST00000155203.1
|
Zbtb10
|
zinc finger and BTB domain containing 10 |
| chr1_-_72536930 | 0.25 |
ENSMUST00000047786.5
|
March4
|
membrane-associated ring finger (C3HC4) 4 |
| chr1_-_119997211 | 0.24 |
ENSMUST00000174458.1
|
Gm101
|
predicted gene 101 |
| chr12_-_69357120 | 0.24 |
ENSMUST00000021368.8
|
Nemf
|
nuclear export mediator factor |
| chr7_+_12881165 | 0.22 |
ENSMUST00000144578.1
|
Zfp128
|
zinc finger protein 128 |
| chr4_+_117096049 | 0.20 |
ENSMUST00000030443.5
|
Ptch2
|
patched homolog 2 |
| chr17_-_84187939 | 0.15 |
ENSMUST00000060366.6
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr12_+_58211772 | 0.15 |
ENSMUST00000110671.2
ENSMUST00000044299.2 |
Sstr1
|
somatostatin receptor 1 |
| chr7_-_133123409 | 0.14 |
ENSMUST00000170459.1
ENSMUST00000166400.1 |
Ctbp2
|
C-terminal binding protein 2 |
| chr2_-_104712122 | 0.14 |
ENSMUST00000111118.1
ENSMUST00000028597.3 |
Tcp11l1
|
t-complex 11 like 1 |
| chr9_-_108578657 | 0.12 |
ENSMUST00000068700.5
|
Wdr6
|
WD repeat domain 6 |
| chr4_-_83486178 | 0.12 |
ENSMUST00000130626.1
|
Psip1
|
PC4 and SFRS1 interacting protein 1 |
| chr9_+_114401076 | 0.11 |
ENSMUST00000063042.9
ENSMUST00000111820.2 |
Glb1
Tmppe
|
galactosidase, beta 1 transmembrane protein with metallophosphoesterase domain |
| chr15_-_77970750 | 0.10 |
ENSMUST00000100484.4
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr7_-_133122615 | 0.08 |
ENSMUST00000167218.1
|
Ctbp2
|
C-terminal binding protein 2 |
| chr10_-_99126321 | 0.07 |
ENSMUST00000060761.5
|
Phxr2
|
per-hexamer repeat gene 2 |
| chr2_-_120563795 | 0.03 |
ENSMUST00000055241.6
ENSMUST00000135625.1 |
Zfp106
|
zinc finger protein 106 |
| chr17_+_68837062 | 0.03 |
ENSMUST00000178545.1
|
Tmem200c
|
transmembrane protein 200C |
| chr5_-_137786651 | 0.02 |
ENSMUST00000031740.9
|
Mepce
|
methylphosphate capping enzyme |
| chr10_+_42502030 | 0.02 |
ENSMUST00000105500.1
ENSMUST00000019939.5 |
Snx3
|
sorting nexin 3 |
| chr4_+_32800246 | 0.01 |
ENSMUST00000062802.4
|
Lyrm2
|
LYR motif containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gli2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.0 | 6.1 | GO:0003383 | apical constriction(GO:0003383) |
| 1.0 | 2.9 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.9 | 2.6 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.8 | 3.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.6 | 2.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.4 | 1.7 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.4 | 1.2 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.4 | 2.4 | GO:2001184 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.4 | 2.3 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.4 | 1.5 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.4 | 4.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.4 | 1.1 | GO:0060365 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) |
| 0.3 | 1.3 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.3 | 0.9 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.3 | 2.8 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 1.0 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.2 | 0.7 | GO:0006296 | base-excision repair, AP site formation(GO:0006285) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.2 | 1.8 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 1.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.2 | 0.9 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.2 | 0.6 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.2 | 1.4 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 0.5 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 1.3 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.2 | 2.7 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.2 | 0.7 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.2 | 1.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.2 | 0.8 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 3.5 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.2 | 1.5 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.2 | 0.6 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 4.1 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 0.8 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 3.8 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 0.6 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 1.8 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 1.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 2.7 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 1.4 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.4 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.1 | 1.0 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 0.5 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.6 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 1.0 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 1.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.1 | 8.8 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 1.6 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 1.0 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 0.3 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 3.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 2.6 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 0.8 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.1 | 0.5 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 1.7 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.5 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 3.8 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 1.1 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 1.0 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 2.0 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.9 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.3 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.4 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 6.4 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.6 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) |
| 0.0 | 0.7 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 1.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 2.7 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.5 | 1.5 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.3 | 3.8 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.3 | 3.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.3 | 8.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 1.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 1.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.5 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.6 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 1.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.2 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 0.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.3 | GO:0090537 | CERF complex(GO:0090537) |
| 0.1 | 0.5 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 2.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.7 | GO:0055038 | melanosome(GO:0042470) pigment granule(GO:0048770) recycling endosome membrane(GO:0055038) |
| 0.0 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.5 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 2.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 2.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 3.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.6 | 2.6 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.5 | 2.8 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.4 | 3.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.4 | 4.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 3.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.4 | 7.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 3.8 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 1.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.3 | 3.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 2.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 1.0 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 1.9 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 2.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.7 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.9 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.7 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 0.5 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 1.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 6.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 2.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.5 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 1.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.5 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.1 | 1.0 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 0.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.2 | GO:0008158 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.6 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.6 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 1.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 4.1 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 2.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.7 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 1.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 2.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.4 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 1.2 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 4.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 2.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 2.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 1.4 | GO:0051219 | phosphoprotein binding(GO:0051219) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 3.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 7.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 3.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 2.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 2.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 3.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 6.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 3.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 2.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.0 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 6.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 0.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.1 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 0.6 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 2.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 2.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |