Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
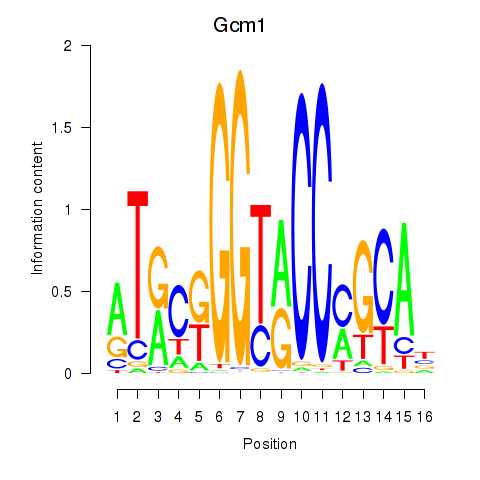
Results for Gcm1
Z-value: 1.20
Transcription factors associated with Gcm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gcm1
|
ENSMUSG00000023333.7 | glial cells missing homolog 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gcm1 | mm10_v2_chr9_+_78051938_78051958 | -0.28 | 9.8e-02 | Click! |
Activity profile of Gcm1 motif
Sorted Z-values of Gcm1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_30373358 | 7.41 |
ENSMUST00000021004.7
|
Sntg2
|
syntrophin, gamma 2 |
| chr15_+_10249560 | 6.29 |
ENSMUST00000134410.1
|
Prlr
|
prolactin receptor |
| chr13_-_4523322 | 5.74 |
ENSMUST00000080361.5
ENSMUST00000078239.3 |
Akr1c20
|
aldo-keto reductase family 1, member C20 |
| chr7_+_119900099 | 4.50 |
ENSMUST00000106516.1
|
Lyrm1
|
LYR motif containing 1 |
| chr3_+_138217814 | 4.34 |
ENSMUST00000090171.5
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr8_-_45358737 | 3.85 |
ENSMUST00000155230.1
ENSMUST00000135912.1 |
Fam149a
|
family with sequence similarity 149, member A |
| chr6_-_124636085 | 3.08 |
ENSMUST00000068797.2
|
Gm5077
|
predicted gene 5077 |
| chr11_-_106613370 | 2.95 |
ENSMUST00000128933.1
|
Tex2
|
testis expressed gene 2 |
| chr17_-_32189457 | 2.90 |
ENSMUST00000087721.3
ENSMUST00000162117.1 |
Ephx3
|
epoxide hydrolase 3 |
| chr8_-_3720640 | 2.65 |
ENSMUST00000062037.6
|
Clec4g
|
C-type lectin domain family 4, member g |
| chr16_+_38902305 | 2.57 |
ENSMUST00000023478.7
|
Igsf11
|
immunoglobulin superfamily, member 11 |
| chr7_-_105600103 | 2.04 |
ENSMUST00000033185.8
|
Hpx
|
hemopexin |
| chr3_+_107230608 | 2.01 |
ENSMUST00000179399.1
|
A630076J17Rik
|
RIKEN cDNA A630076J17 gene |
| chr7_-_99182681 | 1.95 |
ENSMUST00000033001.4
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr3_+_89459325 | 1.94 |
ENSMUST00000107410.1
|
Pmvk
|
phosphomevalonate kinase |
| chr4_-_42661893 | 1.82 |
ENSMUST00000108006.3
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr11_+_120491840 | 1.68 |
ENSMUST00000026899.3
|
Slc25a10
|
solute carrier family 25 (mitochondrial carrier, dicarboxylate transporter), member 10 |
| chr17_-_6948283 | 1.67 |
ENSMUST00000024572.9
|
Rsph3b
|
radial spoke 3B homolog (Chlamydomonas) |
| chr5_+_102724971 | 1.65 |
ENSMUST00000112853.1
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr7_+_131966446 | 1.51 |
ENSMUST00000045840.4
|
Gpr26
|
G protein-coupled receptor 26 |
| chr19_+_46761578 | 1.49 |
ENSMUST00000077666.4
ENSMUST00000099373.4 |
Cnnm2
|
cyclin M2 |
| chr2_+_96318014 | 1.47 |
ENSMUST00000135431.1
ENSMUST00000162807.2 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr2_-_35061431 | 1.44 |
ENSMUST00000028233.3
|
Hc
|
hemolytic complement |
| chr2_-_155834845 | 1.33 |
ENSMUST00000029143.5
|
Fam83c
|
family with sequence similarity 83, member C |
| chr7_-_68264201 | 1.32 |
ENSMUST00000032770.2
|
Pgpep1l
|
pyroglutamyl-peptidase I-like |
| chr11_-_121354337 | 1.26 |
ENSMUST00000106110.3
ENSMUST00000136797.1 ENSMUST00000026173.6 |
Wdr45b
|
WD repeat domain 45B |
| chr7_-_101011976 | 1.25 |
ENSMUST00000178340.1
ENSMUST00000037540.3 |
P2ry2
|
purinergic receptor P2Y, G-protein coupled 2 |
| chr2_+_169633517 | 1.25 |
ENSMUST00000109157.1
|
Tshz2
|
teashirt zinc finger family member 2 |
| chr11_-_35980473 | 1.21 |
ENSMUST00000018993.6
|
Wwc1
|
WW, C2 and coiled-coil domain containing 1 |
| chr14_-_66213552 | 1.19 |
ENSMUST00000178730.1
|
Ptk2b
|
PTK2 protein tyrosine kinase 2 beta |
| chr1_+_174050375 | 1.14 |
ENSMUST00000062665.3
|
Olfr432
|
olfactory receptor 432 |
| chr17_-_24696147 | 1.12 |
ENSMUST00000046839.8
|
Gfer
|
growth factor, erv1 (S. cerevisiae)-like (augmenter of liver regeneration) |
| chr7_+_44997648 | 1.11 |
ENSMUST00000003284.8
ENSMUST00000107835.1 |
Irf3
|
interferon regulatory factor 3 |
| chr11_-_60352869 | 1.09 |
ENSMUST00000095254.5
ENSMUST00000102683.4 ENSMUST00000093048.6 ENSMUST00000093046.6 ENSMUST00000064019.8 ENSMUST00000102682.4 |
Tom1l2
|
target of myb1-like 2 (chicken) |
| chr12_+_8674224 | 1.05 |
ENSMUST00000111122.2
|
Pum2
|
pumilio 2 (Drosophila) |
| chr11_-_98149551 | 1.00 |
ENSMUST00000103143.3
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr9_+_5345414 | 0.98 |
ENSMUST00000027009.4
|
Casp12
|
caspase 12 |
| chr1_+_53313636 | 0.92 |
ENSMUST00000114484.1
|
Osgepl1
|
O-sialoglycoprotein endopeptidase-like 1 |
| chr1_+_53313622 | 0.90 |
ENSMUST00000027265.3
|
Osgepl1
|
O-sialoglycoprotein endopeptidase-like 1 |
| chr17_+_8311101 | 0.86 |
ENSMUST00000154553.1
|
Sft2d1
|
SFT2 domain containing 1 |
| chr7_-_45136391 | 0.78 |
ENSMUST00000146760.1
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr2_+_105904629 | 0.77 |
ENSMUST00000037499.5
|
Immp1l
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr12_+_8674129 | 0.73 |
ENSMUST00000111123.2
ENSMUST00000178015.1 ENSMUST00000020915.3 |
Pum2
|
pumilio 2 (Drosophila) |
| chr12_-_119238794 | 0.73 |
ENSMUST00000026360.8
|
Itgb8
|
integrin beta 8 |
| chr7_-_29906524 | 0.69 |
ENSMUST00000159920.1
ENSMUST00000162592.1 |
Zfp27
|
zinc finger protein 27 |
| chr1_-_191164815 | 0.68 |
ENSMUST00000171798.1
|
Fam71a
|
family with sequence similarity 71, member A |
| chr18_-_39487096 | 0.67 |
ENSMUST00000097592.2
ENSMUST00000115571.1 |
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr13_+_40704005 | 0.67 |
ENSMUST00000069457.1
|
Gm9979
|
predicted gene 9979 |
| chr1_-_79761752 | 0.65 |
ENSMUST00000113512.1
ENSMUST00000113513.1 ENSMUST00000113515.1 ENSMUST00000113514.1 ENSMUST00000113510.1 ENSMUST00000113511.1 ENSMUST00000048820.7 |
Wdfy1
|
WD repeat and FYVE domain containing 1 |
| chr9_+_5345450 | 0.63 |
ENSMUST00000151332.1
|
Casp12
|
caspase 12 |
| chr16_+_36875119 | 0.62 |
ENSMUST00000135406.1
ENSMUST00000114812.1 ENSMUST00000134616.1 ENSMUST00000023534.6 |
Golgb1
Golgb1
|
golgi autoantigen, golgin subfamily b, macrogolgin 1 golgi autoantigen, golgin subfamily b, macrogolgin 1 |
| chr8_+_91070052 | 0.61 |
ENSMUST00000034091.7
|
Rbl2
|
retinoblastoma-like 2 |
| chr9_+_107580117 | 0.61 |
ENSMUST00000093785.4
|
Nat6
|
N-acetyltransferase 6 |
| chr8_+_105170668 | 0.60 |
ENSMUST00000109395.1
ENSMUST00000109394.1 ENSMUST00000052209.2 ENSMUST00000109392.1 |
Cbfb
|
core binding factor beta |
| chr7_-_45136102 | 0.59 |
ENSMUST00000125500.1
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr4_+_155451570 | 0.58 |
ENSMUST00000135407.1
ENSMUST00000105619.1 |
C030017K20Rik
|
RIKEN cDNA C030017K20 gene |
| chr15_-_10470490 | 0.57 |
ENSMUST00000136591.1
|
Dnajc21
|
DnaJ (Hsp40) homolog, subfamily C, member 21 |
| chr2_-_105904484 | 0.56 |
ENSMUST00000122965.1
|
Elp4
|
elongation protein 4 homolog (S. cerevisiae) |
| chr14_-_32201869 | 0.55 |
ENSMUST00000170331.1
ENSMUST00000013845.6 |
Timm23
|
translocase of inner mitochondrial membrane 23 |
| chr6_+_86365673 | 0.54 |
ENSMUST00000071492.7
|
Fam136a
|
family with sequence similarity 136, member A |
| chr15_+_85735564 | 0.54 |
ENSMUST00000109423.1
|
Ppara
|
peroxisome proliferator activated receptor alpha |
| chr11_-_75454656 | 0.53 |
ENSMUST00000173320.1
|
Wdr81
|
WD repeat domain 81 |
| chr13_+_100108155 | 0.52 |
ENSMUST00000129014.1
|
Serf1
|
small EDRK-rich factor 1 |
| chr9_-_107605295 | 0.51 |
ENSMUST00000102529.3
|
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr7_+_46396439 | 0.51 |
ENSMUST00000025202.6
|
Kcnc1
|
potassium voltage gated channel, Shaw-related subfamily, member 1 |
| chr10_-_71285234 | 0.48 |
ENSMUST00000020085.6
|
Ube2d1
|
ubiquitin-conjugating enzyme E2D 1 |
| chr6_-_42461017 | 0.46 |
ENSMUST00000090156.1
|
Olfr458
|
olfactory receptor 458 |
| chr10_+_80148263 | 0.42 |
ENSMUST00000099492.3
ENSMUST00000042057.5 |
Midn
|
midnolin |
| chr7_-_45136056 | 0.42 |
ENSMUST00000130628.1
|
Flt3l
|
FMS-like tyrosine kinase 3 ligand |
| chr13_-_95250166 | 0.41 |
ENSMUST00000162153.1
ENSMUST00000160957.2 ENSMUST00000159598.1 ENSMUST00000162412.1 |
Pde8b
|
phosphodiesterase 8B |
| chr11_-_6065538 | 0.40 |
ENSMUST00000101585.3
ENSMUST00000066431.7 ENSMUST00000109815.2 ENSMUST00000109812.2 ENSMUST00000101586.2 ENSMUST00000093355.5 ENSMUST00000019133.4 |
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta |
| chr1_+_43092588 | 0.39 |
ENSMUST00000039080.3
|
8430432A02Rik
|
RIKEN cDNA 8430432A02 gene |
| chr11_-_73199013 | 0.37 |
ENSMUST00000006103.2
ENSMUST00000108476.1 |
Ctns
|
cystinosis, nephropathic |
| chr7_+_29519158 | 0.32 |
ENSMUST00000141713.1
|
4932431P20Rik
|
RIKEN cDNA 4932431P20 gene |
| chrX_+_73503074 | 0.30 |
ENSMUST00000114479.1
ENSMUST00000088429.1 ENSMUST00000033744.5 |
Atp2b3
|
ATPase, Ca++ transporting, plasma membrane 3 |
| chr2_+_181864337 | 0.29 |
ENSMUST00000039551.8
|
Polr3k
|
polymerase (RNA) III (DNA directed) polypeptide K |
| chr11_+_116918844 | 0.28 |
ENSMUST00000103027.3
|
Mgat5b
|
mannoside acetylglucosaminyltransferase 5, isoenzyme B |
| chr13_+_100107997 | 0.28 |
ENSMUST00000142155.1
ENSMUST00000022145.8 ENSMUST00000132053.1 |
Serf1
|
small EDRK-rich factor 1 |
| chr8_-_13254096 | 0.25 |
ENSMUST00000171619.1
|
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr10_+_52388963 | 0.25 |
ENSMUST00000160539.1
|
Nepn
|
nephrocan |
| chr15_-_83251720 | 0.24 |
ENSMUST00000164614.1
ENSMUST00000049530.6 |
A4galt
|
alpha 1,4-galactosyltransferase |
| chr17_+_24696234 | 0.22 |
ENSMUST00000019464.7
|
Noxo1
|
NADPH oxidase organizer 1 |
| chr16_-_36874806 | 0.15 |
ENSMUST00000075946.5
|
Eaf2
|
ELL associated factor 2 |
| chr8_-_13254068 | 0.14 |
ENSMUST00000168498.1
|
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr2_+_48814109 | 0.13 |
ENSMUST00000063886.3
|
Acvr2a
|
activin receptor IIA |
| chr13_+_8202922 | 0.12 |
ENSMUST00000123187.1
|
Adarb2
|
adenosine deaminase, RNA-specific, B2 |
| chr13_-_100108337 | 0.11 |
ENSMUST00000180822.1
|
BC001981
|
cDNA sequence BC001981 |
| chr4_-_156234777 | 0.10 |
ENSMUST00000105569.3
|
Klhl17
|
kelch-like 17 |
| chr13_+_8202860 | 0.08 |
ENSMUST00000064473.6
|
Adarb2
|
adenosine deaminase, RNA-specific, B2 |
| chrX_+_7657260 | 0.08 |
ENSMUST00000033485.7
|
Prickle3
|
prickle homolog 3 (Drosophila) |
| chr17_-_32788284 | 0.06 |
ENSMUST00000159086.2
|
Zfp871
|
zinc finger protein 871 |
| chr6_+_6863769 | 0.04 |
ENSMUST00000031768.6
|
Dlx6
|
distal-less homeobox 6 |
| chr7_+_4922251 | 0.03 |
ENSMUST00000047309.5
|
Nat14
|
N-acetyltransferase 14 |
| chrX_+_93286499 | 0.00 |
ENSMUST00000046565.7
ENSMUST00000113947.2 |
Arx
|
aristaless related homeobox |
| chr7_-_44997535 | 0.00 |
ENSMUST00000124232.1
ENSMUST00000003290.4 |
Bcl2l12
|
BCL2-like 12 (proline rich) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gcm1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 1.0 | 6.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.5 | 1.9 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.5 | 1.8 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.4 | 1.7 | GO:0015744 | succinate transport(GO:0015744) |
| 0.4 | 2.0 | GO:0015886 | heme transport(GO:0015886) |
| 0.4 | 1.2 | GO:0007172 | signal complex assembly(GO:0007172) regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.3 | 1.4 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.3 | 1.1 | GO:0045358 | negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.2 | 2.0 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.2 | 1.6 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 1.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 1.8 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 0.4 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 0.1 | 0.7 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 | 2.7 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 1.3 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.1 | 0.4 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 1.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.8 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.5 | GO:0072363 | regulation of glycolytic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072363) |
| 0.1 | 1.1 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.1 | 0.5 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 1.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.4 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 1.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.3 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.6 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.1 | 0.4 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.5 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.2 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.0 | 0.7 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0046881 | sperm ejaculation(GO:0042713) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.5 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 5.7 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 1.5 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 1.1 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 1.5 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 1.1 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 1.0 | GO:0001662 | behavioral fear response(GO:0001662) |
| 0.0 | 0.3 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.6 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.3 | 0.8 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.6 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 1.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 4.5 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.5 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 2.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.2 | GO:0032587 | ruffle membrane(GO:0032587) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 1.3 | 6.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.7 | 2.0 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.6 | 1.8 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.6 | 7.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.6 | 1.7 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.5 | 2.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.4 | 1.3 | GO:0031686 | uridine nucleotide receptor activity(GO:0015065) A1 adenosine receptor binding(GO:0031686) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.3 | 1.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 5.7 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 1.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.7 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.4 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 1.6 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 0.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 2.7 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.6 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.3 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 1.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.6 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 1.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 1.9 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 1.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 1.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 5.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 6.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 1.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.0 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |