Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
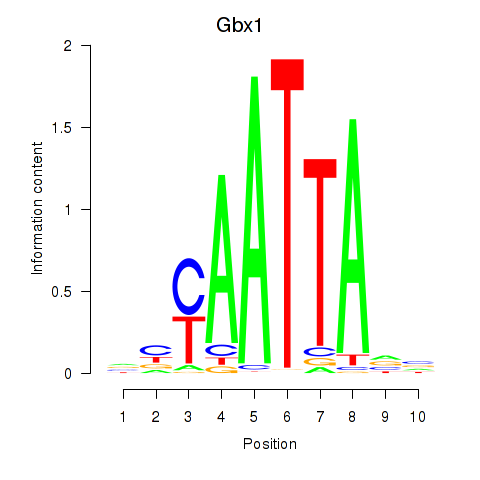
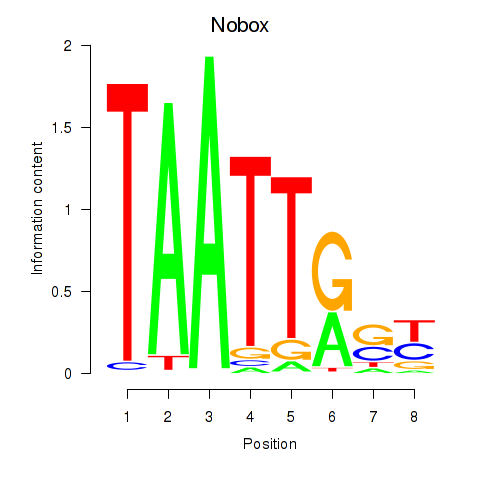
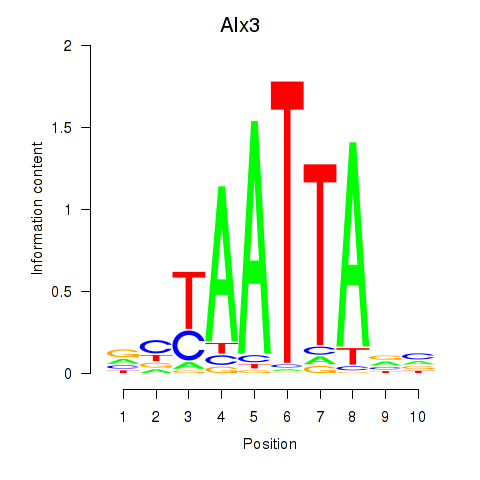
Results for Gbx1_Nobox_Alx3
Z-value: 1.06
Transcription factors associated with Gbx1_Nobox_Alx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gbx1
|
ENSMUSG00000067724.4 | gastrulation brain homeobox 1 |
|
Nobox
|
ENSMUSG00000029736.9 | NOBOX oogenesis homeobox |
|
Alx3
|
ENSMUSG00000014603.1 | aristaless-like homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Alx3 | mm10_v2_chr3_+_107595031_107595164 | -0.22 | 1.9e-01 | Click! |
| Gbx1 | mm10_v2_chr5_-_24527276_24527276 | 0.04 | 8.2e-01 | Click! |
| Nobox | mm10_v2_chr6_-_43307236_43307236 | -0.02 | 9.2e-01 | Click! |
Activity profile of Gbx1_Nobox_Alx3 motif
Sorted Z-values of Gbx1_Nobox_Alx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_99322943 | 7.06 |
ENSMUST00000038004.2
|
Krt25
|
keratin 25 |
| chr19_+_40089688 | 6.44 |
ENSMUST00000068094.6
ENSMUST00000080171.2 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr19_-_7966000 | 6.20 |
ENSMUST00000182102.1
ENSMUST00000075619.4 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr13_+_4434306 | 5.93 |
ENSMUST00000021630.8
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr10_+_127898515 | 5.44 |
ENSMUST00000047134.7
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr6_+_37870786 | 2.62 |
ENSMUST00000120428.1
ENSMUST00000031859.7 |
Trim24
|
tripartite motif-containing 24 |
| chr14_+_33941021 | 2.60 |
ENSMUST00000100720.1
|
Gdf2
|
growth differentiation factor 2 |
| chr9_-_71163224 | 2.58 |
ENSMUST00000074465.2
|
Aqp9
|
aquaporin 9 |
| chr3_+_62419668 | 2.31 |
ENSMUST00000161057.1
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr19_+_26749726 | 2.20 |
ENSMUST00000175842.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr8_-_61902669 | 2.00 |
ENSMUST00000121785.1
ENSMUST00000034057.7 |
Palld
|
palladin, cytoskeletal associated protein |
| chr4_+_108719649 | 1.91 |
ENSMUST00000178992.1
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr1_-_72284248 | 1.90 |
ENSMUST00000097698.4
ENSMUST00000027381.6 |
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr7_+_126950687 | 1.82 |
ENSMUST00000106333.1
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr2_+_20737306 | 1.77 |
ENSMUST00000114606.1
ENSMUST00000114608.1 |
Etl4
|
enhancer trap locus 4 |
| chr7_+_126950518 | 1.74 |
ENSMUST00000106335.1
ENSMUST00000146017.1 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chr8_-_25038875 | 1.73 |
ENSMUST00000084031.4
|
Htra4
|
HtrA serine peptidase 4 |
| chr3_-_49757257 | 1.72 |
ENSMUST00000035931.7
|
Pcdh18
|
protocadherin 18 |
| chr5_-_62766153 | 1.65 |
ENSMUST00000076623.4
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr5_-_134747241 | 1.51 |
ENSMUST00000015138.9
|
Eln
|
elastin |
| chr12_-_91849081 | 1.48 |
ENSMUST00000167466.1
ENSMUST00000021347.5 ENSMUST00000178462.1 |
Sel1l
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr5_-_62765618 | 1.46 |
ENSMUST00000159470.1
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr18_-_75697639 | 1.45 |
ENSMUST00000165559.1
|
Ctif
|
CBP80/20-dependent translation initiation factor |
| chr11_-_99374895 | 1.41 |
ENSMUST00000006963.2
|
Krt28
|
keratin 28 |
| chrM_+_10167 | 1.39 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr15_-_101892916 | 1.36 |
ENSMUST00000100179.1
|
Krt76
|
keratin 76 |
| chr8_+_45658666 | 1.34 |
ENSMUST00000134675.1
ENSMUST00000139869.1 ENSMUST00000126067.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr16_+_22918378 | 1.34 |
ENSMUST00000170805.1
|
Fetub
|
fetuin beta |
| chr2_+_109917639 | 1.33 |
ENSMUST00000046548.7
ENSMUST00000111037.2 |
Lgr4
|
leucine-rich repeat-containing G protein-coupled receptor 4 |
| chr1_-_190170671 | 1.31 |
ENSMUST00000175916.1
|
Prox1
|
prospero-related homeobox 1 |
| chr1_+_72284367 | 1.28 |
ENSMUST00000027380.5
ENSMUST00000141783.1 |
Tmem169
|
transmembrane protein 169 |
| chr5_+_90561102 | 1.28 |
ENSMUST00000094615.4
|
5830473C10Rik
|
RIKEN cDNA 5830473C10 gene |
| chrM_+_9870 | 1.26 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr7_+_103550368 | 1.25 |
ENSMUST00000106888.1
|
Olfr613
|
olfactory receptor 613 |
| chr1_+_132298606 | 1.25 |
ENSMUST00000046071.4
|
Klhdc8a
|
kelch domain containing 8A |
| chr4_+_34893772 | 1.25 |
ENSMUST00000029975.3
ENSMUST00000135871.1 ENSMUST00000108130.1 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr15_-_37459327 | 1.24 |
ENSMUST00000119730.1
ENSMUST00000120746.1 |
Ncald
|
neurocalcin delta |
| chr19_+_39992424 | 1.24 |
ENSMUST00000049178.2
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr5_+_139543889 | 1.23 |
ENSMUST00000174792.1
ENSMUST00000031523.8 |
Uncx
|
UNC homeobox |
| chr7_+_79273201 | 1.20 |
ENSMUST00000037315.6
|
Abhd2
|
abhydrolase domain containing 2 |
| chr10_-_107486077 | 1.19 |
ENSMUST00000000445.1
|
Myf5
|
myogenic factor 5 |
| chr11_-_99244058 | 1.17 |
ENSMUST00000103132.3
ENSMUST00000038214.6 |
Krt222
|
keratin 222 |
| chr15_+_55307743 | 1.10 |
ENSMUST00000023053.5
ENSMUST00000110221.2 ENSMUST00000110217.3 |
Col14a1
|
collagen, type XIV, alpha 1 |
| chr7_+_64501687 | 1.06 |
ENSMUST00000032732.8
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr18_+_56432116 | 1.05 |
ENSMUST00000070166.5
|
Gramd3
|
GRAM domain containing 3 |
| chr2_-_37359274 | 1.02 |
ENSMUST00000009174.8
|
Pdcl
|
phosducin-like |
| chr1_+_161494649 | 1.02 |
ENSMUST00000086084.1
|
Tnfsf18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr18_+_84851338 | 0.98 |
ENSMUST00000160180.1
|
Cyb5
|
cytochrome b-5 |
| chr17_-_83846769 | 0.97 |
ENSMUST00000000687.7
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr2_+_23069210 | 0.95 |
ENSMUST00000155602.1
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr3_-_79841729 | 0.94 |
ENSMUST00000168038.1
|
Tmem144
|
transmembrane protein 144 |
| chr8_+_45658731 | 0.92 |
ENSMUST00000143820.1
ENSMUST00000132139.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr5_+_9100681 | 0.91 |
ENSMUST00000115365.1
|
Tmem243
|
transmembrane protein 243, mitochondrial |
| chrX_-_60403947 | 0.89 |
ENSMUST00000033480.6
ENSMUST00000101527.2 |
Atp11c
|
ATPase, class VI, type 11C |
| chr17_+_45734506 | 0.87 |
ENSMUST00000180558.1
|
F630040K05Rik
|
RIKEN cDNA F630040K05 gene |
| chr1_+_74284930 | 0.87 |
ENSMUST00000113805.1
ENSMUST00000027370.6 ENSMUST00000087226.4 |
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr7_-_105600103 | 0.87 |
ENSMUST00000033185.8
|
Hpx
|
hemopexin |
| chr12_+_111814170 | 0.86 |
ENSMUST00000021714.7
|
Zfyve21
|
zinc finger, FYVE domain containing 21 |
| chr4_-_129261394 | 0.84 |
ENSMUST00000145261.1
|
C77080
|
expressed sequence C77080 |
| chr3_-_32491969 | 0.83 |
ENSMUST00000164954.1
|
Kcnmb3
|
potassium large conductance calcium-activated channel, subfamily M, beta member 3 |
| chr1_-_121332545 | 0.82 |
ENSMUST00000161068.1
|
Insig2
|
insulin induced gene 2 |
| chr10_+_39612934 | 0.82 |
ENSMUST00000019987.6
|
Traf3ip2
|
TRAF3 interacting protein 2 |
| chrX_-_143933089 | 0.82 |
ENSMUST00000087313.3
|
Dcx
|
doublecortin |
| chr8_+_45627709 | 0.81 |
ENSMUST00000134321.1
ENSMUST00000135336.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chrM_+_7759 | 0.79 |
ENSMUST00000082407.1
ENSMUST00000082408.1 |
mt-Atp8
mt-Atp6
|
mitochondrially encoded ATP synthase 8 mitochondrially encoded ATP synthase 6 |
| chr16_+_45094036 | 0.78 |
ENSMUST00000061050.5
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr3_+_122419772 | 0.78 |
ENSMUST00000029766.4
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr11_-_20332654 | 0.77 |
ENSMUST00000004634.6
|
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr11_-_20332689 | 0.76 |
ENSMUST00000109594.1
|
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr15_-_37458523 | 0.76 |
ENSMUST00000116445.2
|
Ncald
|
neurocalcin delta |
| chr6_-_130231638 | 0.75 |
ENSMUST00000088011.4
ENSMUST00000112013.1 ENSMUST00000049304.7 |
Klra7
|
killer cell lectin-like receptor, subfamily A, member 7 |
| chr5_-_137684665 | 0.74 |
ENSMUST00000100544.4
ENSMUST00000031736.9 ENSMUST00000151839.1 |
Agfg2
|
ArfGAP with FG repeats 2 |
| chrM_+_7005 | 0.73 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr5_+_135106881 | 0.73 |
ENSMUST00000005507.3
|
Mlxipl
|
MLX interacting protein-like |
| chr12_-_56613270 | 0.73 |
ENSMUST00000072631.5
|
Nkx2-9
|
NK2 homeobox 9 |
| chr2_-_116067391 | 0.73 |
ENSMUST00000140185.1
|
2700033N17Rik
|
RIKEN cDNA 2700033N17 gene |
| chr11_-_107337556 | 0.73 |
ENSMUST00000040380.6
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr13_-_53473074 | 0.73 |
ENSMUST00000021922.8
|
Msx2
|
msh homeobox 2 |
| chr2_-_77519565 | 0.72 |
ENSMUST00000111830.2
|
Zfp385b
|
zinc finger protein 385B |
| chr11_+_99873389 | 0.72 |
ENSMUST00000093936.3
|
Krtap9-1
|
keratin associated protein 9-1 |
| chr3_+_121291725 | 0.71 |
ENSMUST00000039442.7
|
Alg14
|
asparagine-linked glycosylation 14 |
| chr8_-_45382198 | 0.70 |
ENSMUST00000093526.6
|
Fam149a
|
family with sequence similarity 149, member A |
| chr16_+_37580137 | 0.69 |
ENSMUST00000160847.1
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chrX_-_60893430 | 0.69 |
ENSMUST00000135107.2
|
Sox3
|
SRY-box containing gene 3 |
| chr1_-_24612700 | 0.68 |
ENSMUST00000088336.1
|
Gm10222
|
predicted gene 10222 |
| chr16_-_63864114 | 0.68 |
ENSMUST00000064405.6
|
Epha3
|
Eph receptor A3 |
| chr5_-_87482258 | 0.68 |
ENSMUST00000079811.6
ENSMUST00000144144.1 |
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr2_+_69897220 | 0.68 |
ENSMUST00000055758.9
ENSMUST00000112251.2 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr19_+_26750939 | 0.67 |
ENSMUST00000175953.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_-_45103747 | 0.67 |
ENSMUST00000003512.7
|
Fcgrt
|
Fc receptor, IgG, alpha chain transporter |
| chr6_-_101377897 | 0.67 |
ENSMUST00000075994.6
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr2_+_69897255 | 0.66 |
ENSMUST00000131553.1
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr9_-_103222063 | 0.65 |
ENSMUST00000170904.1
|
Trf
|
transferrin |
| chr9_+_43829963 | 0.64 |
ENSMUST00000180221.1
|
Gm3898
|
predicted gene 3898 |
| chr17_-_59013264 | 0.63 |
ENSMUST00000174122.1
ENSMUST00000025065.5 |
Nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr6_-_57535422 | 0.62 |
ENSMUST00000042766.3
|
Ppm1k
|
protein phosphatase 1K (PP2C domain containing) |
| chr12_-_57546121 | 0.62 |
ENSMUST00000044380.6
|
Foxa1
|
forkhead box A1 |
| chr13_-_102905740 | 0.62 |
ENSMUST00000167462.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr15_+_25773985 | 0.62 |
ENSMUST00000125667.1
|
Myo10
|
myosin X |
| chr1_-_121332571 | 0.61 |
ENSMUST00000071064.6
|
Insig2
|
insulin induced gene 2 |
| chr2_+_86046451 | 0.61 |
ENSMUST00000079298.2
|
Olfr1034
|
olfactory receptor 1034 |
| chr11_+_94327984 | 0.61 |
ENSMUST00000107818.2
ENSMUST00000051221.6 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr13_-_85127514 | 0.59 |
ENSMUST00000179230.1
|
Gm4076
|
predicted gene 4076 |
| chr2_-_27475622 | 0.59 |
ENSMUST00000138693.1
ENSMUST00000113941.2 ENSMUST00000077737.6 |
Brd3
|
bromodomain containing 3 |
| chr2_+_155382186 | 0.58 |
ENSMUST00000134218.1
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr10_+_127421208 | 0.58 |
ENSMUST00000168780.1
|
R3hdm2
|
R3H domain containing 2 |
| chr10_-_92375367 | 0.58 |
ENSMUST00000182870.1
|
Gm20757
|
predicted gene, 20757 |
| chr13_+_42680565 | 0.58 |
ENSMUST00000128646.1
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr2_+_128591205 | 0.58 |
ENSMUST00000155430.1
|
Gm355
|
predicted gene 355 |
| chr1_-_152625212 | 0.57 |
ENSMUST00000027760.7
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr11_+_87663087 | 0.57 |
ENSMUST00000165679.1
|
Rnf43
|
ring finger protein 43 |
| chr7_-_99141068 | 0.57 |
ENSMUST00000037968.8
|
Uvrag
|
UV radiation resistance associated gene |
| chr4_+_144893127 | 0.55 |
ENSMUST00000142808.1
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr3_+_57425314 | 0.55 |
ENSMUST00000029377.7
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr2_+_116067213 | 0.55 |
ENSMUST00000152412.1
|
G630016G05Rik
|
RIKEN cDNA G630016G05 gene |
| chr7_-_116198487 | 0.55 |
ENSMUST00000181981.1
|
Plekha7
|
pleckstrin homology domain containing, family A member 7 |
| chr11_-_99521258 | 0.54 |
ENSMUST00000076948.1
|
Krt39
|
keratin 39 |
| chr18_-_3337467 | 0.54 |
ENSMUST00000154135.1
|
Crem
|
cAMP responsive element modulator |
| chr2_-_64975762 | 0.54 |
ENSMUST00000156765.1
|
Grb14
|
growth factor receptor bound protein 14 |
| chr14_+_73237891 | 0.54 |
ENSMUST00000044405.6
|
Lpar6
|
lysophosphatidic acid receptor 6 |
| chr16_+_33684538 | 0.53 |
ENSMUST00000126532.1
|
Heg1
|
HEG homolog 1 (zebrafish) |
| chr5_+_88583527 | 0.53 |
ENSMUST00000031229.6
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr6_-_115592571 | 0.53 |
ENSMUST00000112957.1
|
2510049J12Rik
|
RIKEN cDNA 2510049J12 gene |
| chr3_+_68572245 | 0.52 |
ENSMUST00000170788.2
|
Schip1
|
schwannomin interacting protein 1 |
| chr2_-_10130638 | 0.52 |
ENSMUST00000042290.7
|
Itih2
|
inter-alpha trypsin inhibitor, heavy chain 2 |
| chr3_+_55461758 | 0.52 |
ENSMUST00000070418.4
|
Dclk1
|
doublecortin-like kinase 1 |
| chr6_-_116716888 | 0.52 |
ENSMUST00000056623.6
|
Tmem72
|
transmembrane protein 72 |
| chr9_+_32116040 | 0.52 |
ENSMUST00000174641.1
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr1_-_190170178 | 0.51 |
ENSMUST00000177288.1
|
Prox1
|
prospero-related homeobox 1 |
| chr4_+_150853919 | 0.51 |
ENSMUST00000073600.2
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr13_+_94083490 | 0.51 |
ENSMUST00000156071.1
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chr16_+_13358375 | 0.50 |
ENSMUST00000149359.1
|
Mkl2
|
MKL/myocardin-like 2 |
| chr5_+_42067960 | 0.50 |
ENSMUST00000087332.4
|
Gm16223
|
predicted gene 16223 |
| chr7_+_64502090 | 0.50 |
ENSMUST00000137732.1
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr9_+_45138437 | 0.50 |
ENSMUST00000060125.5
|
Scn4b
|
sodium channel, type IV, beta |
| chr2_-_71750083 | 0.49 |
ENSMUST00000180494.1
|
Gm17250
|
predicted gene, 17250 |
| chr10_+_127420334 | 0.48 |
ENSMUST00000171434.1
|
R3hdm2
|
R3H domain containing 2 |
| chrX_+_160768179 | 0.48 |
ENSMUST00000112368.2
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr1_+_177729814 | 0.47 |
ENSMUST00000016106.5
|
1700016C15Rik
|
RIKEN cDNA 1700016C15 gene |
| chr14_-_18893623 | 0.47 |
ENSMUST00000177259.1
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr5_+_29195983 | 0.47 |
ENSMUST00000160888.1
ENSMUST00000159272.1 ENSMUST00000001247.5 ENSMUST00000161398.1 ENSMUST00000160246.1 |
Rnf32
|
ring finger protein 32 |
| chr3_+_5218546 | 0.47 |
ENSMUST00000026284.6
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr1_-_4360256 | 0.46 |
ENSMUST00000027032.4
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr3_+_5218516 | 0.46 |
ENSMUST00000175866.1
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr14_-_36919314 | 0.46 |
ENSMUST00000182797.1
|
Ccser2
|
coiled-coil serine rich 2 |
| chr6_-_126645784 | 0.46 |
ENSMUST00000055168.3
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 |
| chr6_-_136171722 | 0.45 |
ENSMUST00000053880.6
|
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr1_+_82233112 | 0.43 |
ENSMUST00000023262.5
|
Gm9747
|
predicted gene 9747 |
| chr7_+_126950837 | 0.43 |
ENSMUST00000106332.1
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr16_-_37384915 | 0.43 |
ENSMUST00000114787.1
ENSMUST00000114782.1 ENSMUST00000023526.2 ENSMUST00000114775.1 |
Stxbp5l
|
syntaxin binding protein 5-like |
| chr4_-_63154130 | 0.43 |
ENSMUST00000030041.4
|
Ambp
|
alpha 1 microglobulin/bikunin |
| chr15_-_11037968 | 0.43 |
ENSMUST00000058007.5
|
Rxfp3
|
relaxin family peptide receptor 3 |
| chr17_+_45506825 | 0.43 |
ENSMUST00000024733.7
|
Aars2
|
alanyl-tRNA synthetase 2, mitochondrial (putative) |
| chr15_-_33687840 | 0.43 |
ENSMUST00000042021.3
|
Tspyl5
|
testis-specific protein, Y-encoded-like 5 |
| chr1_-_155417394 | 0.42 |
ENSMUST00000111775.1
ENSMUST00000111774.1 |
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr3_+_86070915 | 0.42 |
ENSMUST00000182666.1
|
Sh3d19
|
SH3 domain protein D19 |
| chr10_+_127421124 | 0.42 |
ENSMUST00000170336.1
|
R3hdm2
|
R3H domain containing 2 |
| chr18_-_3337614 | 0.41 |
ENSMUST00000150235.1
ENSMUST00000154470.1 |
Crem
|
cAMP responsive element modulator |
| chr18_-_3281036 | 0.41 |
ENSMUST00000049942.6
ENSMUST00000139537.1 ENSMUST00000124747.1 |
Crem
|
cAMP responsive element modulator |
| chr14_-_18893376 | 0.41 |
ENSMUST00000151926.1
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chrX_+_107255878 | 0.40 |
ENSMUST00000101294.2
ENSMUST00000118820.1 ENSMUST00000120971.1 |
Gpr174
|
G protein-coupled receptor 174 |
| chr11_+_70166623 | 0.40 |
ENSMUST00000102571.3
ENSMUST00000178945.1 ENSMUST00000000327.6 |
Clec10a
|
C-type lectin domain family 10, member A |
| chr8_-_84846860 | 0.40 |
ENSMUST00000003912.6
|
Calr
|
calreticulin |
| chr17_-_49564262 | 0.40 |
ENSMUST00000057610.6
|
Daam2
|
dishevelled associated activator of morphogenesis 2 |
| chr17_-_35697971 | 0.40 |
ENSMUST00000146472.1
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr2_-_134554348 | 0.39 |
ENSMUST00000028704.2
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr6_+_8948608 | 0.38 |
ENSMUST00000160300.1
|
Nxph1
|
neurexophilin 1 |
| chr5_+_81021202 | 0.38 |
ENSMUST00000117253.1
ENSMUST00000120128.1 |
Lphn3
|
latrophilin 3 |
| chr18_-_3337539 | 0.38 |
ENSMUST00000142690.1
ENSMUST00000025069.4 ENSMUST00000082141.5 ENSMUST00000165086.1 ENSMUST00000149803.1 |
Crem
|
cAMP responsive element modulator |
| chr3_+_121967822 | 0.38 |
ENSMUST00000137089.1
|
Arhgap29
|
Rho GTPase activating protein 29 |
| chr11_-_99521336 | 0.37 |
ENSMUST00000107445.1
|
Krt39
|
keratin 39 |
| chr15_-_67113909 | 0.37 |
ENSMUST00000092640.5
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr6_+_15196949 | 0.37 |
ENSMUST00000151301.1
ENSMUST00000131414.1 ENSMUST00000140557.1 ENSMUST00000115469.1 |
Foxp2
|
forkhead box P2 |
| chr8_-_67818218 | 0.36 |
ENSMUST00000059374.4
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr10_+_73821937 | 0.35 |
ENSMUST00000134009.2
ENSMUST00000125006.2 ENSMUST00000177420.1 |
Pcdh15
|
protocadherin 15 |
| chr11_+_29718563 | 0.35 |
ENSMUST00000060992.5
|
Rtn4
|
reticulon 4 |
| chr8_-_67818284 | 0.34 |
ENSMUST00000120071.1
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr11_+_23306884 | 0.34 |
ENSMUST00000180046.1
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr6_+_145934113 | 0.34 |
ENSMUST00000032383.7
|
Sspn
|
sarcospan |
| chr6_-_147264124 | 0.34 |
ENSMUST00000052296.6
|
Pthlh
|
parathyroid hormone-like peptide |
| chr18_-_38284391 | 0.34 |
ENSMUST00000025311.5
|
Pcdh12
|
protocadherin 12 |
| chr1_-_172632931 | 0.33 |
ENSMUST00000027826.5
|
Dusp23
|
dual specificity phosphatase 23 |
| chr6_-_148831395 | 0.33 |
ENSMUST00000145960.1
|
Ipo8
|
importin 8 |
| chr6_+_34746368 | 0.33 |
ENSMUST00000142716.1
|
Cald1
|
caldesmon 1 |
| chr11_-_43901187 | 0.32 |
ENSMUST00000067258.2
ENSMUST00000139906.1 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr12_-_83921809 | 0.32 |
ENSMUST00000135962.1
ENSMUST00000155112.1 ENSMUST00000136848.1 ENSMUST00000126943.1 |
Numb
|
numb gene homolog (Drosophila) |
| chr7_+_125829653 | 0.32 |
ENSMUST00000124223.1
|
D430042O09Rik
|
RIKEN cDNA D430042O09 gene |
| chr11_+_94328242 | 0.32 |
ENSMUST00000021227.5
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr11_+_70166696 | 0.32 |
ENSMUST00000178567.1
|
Clec10a
|
C-type lectin domain family 10, member A |
| chr11_-_99337930 | 0.32 |
ENSMUST00000100482.2
|
Krt26
|
keratin 26 |
| chr1_+_104768510 | 0.32 |
ENSMUST00000062528.8
|
Cdh20
|
cadherin 20 |
| chr8_-_84662841 | 0.31 |
ENSMUST00000060427.4
|
Ier2
|
immediate early response 2 |
| chr4_-_14621669 | 0.31 |
ENSMUST00000143105.1
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr7_+_19228334 | 0.31 |
ENSMUST00000063976.8
|
Opa3
|
optic atrophy 3 |
| chrM_+_2743 | 0.30 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr1_-_92518515 | 0.30 |
ENSMUST00000062353.4
|
Olfr1414
|
olfactory receptor 1414 |
| chr9_-_67539392 | 0.30 |
ENSMUST00000039662.8
|
Tln2
|
talin 2 |
| chrX_-_143933204 | 0.30 |
ENSMUST00000112851.1
ENSMUST00000112856.2 ENSMUST00000033642.3 |
Dcx
|
doublecortin |
| chr3_+_93393696 | 0.30 |
ENSMUST00000045912.2
|
Rptn
|
repetin |
| chr7_+_30458280 | 0.30 |
ENSMUST00000126297.1
|
Nphs1
|
nephrosis 1, nephrin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gbx1_Nobox_Alx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.6 | 2.6 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.6 | 1.8 | GO:0061114 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.6 | 6.4 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.4 | 1.3 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.4 | 6.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.4 | 1.5 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.4 | 2.6 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.3 | 1.0 | GO:1902605 | heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.3 | 1.0 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.3 | 0.3 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.3 | 1.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.3 | 2.9 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.2 | 0.7 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 0.2 | 0.7 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.2 | 0.6 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.2 | 0.6 | GO:0030862 | neuroblast division in subventricular zone(GO:0021849) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.2 | 0.5 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.2 | 0.9 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 6.7 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.2 | 1.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 0.5 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.2 | 0.6 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 0.6 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.2 | 1.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.6 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.7 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 2.6 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 4.0 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 0.4 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 1.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.5 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.4 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.1 | 1.4 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 1.9 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 1.4 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.7 | GO:0009757 | carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.1 | 1.0 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.1 | 0.7 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) |
| 0.1 | 0.2 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.1 | 0.5 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.5 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.9 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 0.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.3 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 1.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 2.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.5 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.1 | 0.4 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 1.3 | GO:0006590 | thyroid hormone generation(GO:0006590) follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 0.3 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.1 | 0.6 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 1.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.6 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.1 | 2.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.4 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.1 | 0.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.2 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 1.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.2 | GO:0090172 | microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 0.1 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.7 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.3 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 3.0 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 1.5 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0061325 | cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.0 | 0.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 2.0 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 1.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.7 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.9 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.5 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.5 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 1.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:1903140 | regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.0 | 0.8 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.9 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 1.9 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.8 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.7 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 1.2 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.5 | GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) |
| 0.0 | 0.5 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.1 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 0.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.2 | GO:2000121 | elastic fiber assembly(GO:0048251) regulation of removal of superoxide radicals(GO:2000121) |
| 0.0 | 0.1 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 0.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.2 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.0 | GO:0060618 | nipple development(GO:0060618) |
| 0.0 | 0.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.3 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.2 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 1.5 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.2 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.4 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.0 | GO:0002587 | negative regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002587) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.3 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.5 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.4 | 2.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.3 | 1.5 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 1.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 2.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 11.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.0 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 1.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.7 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.4 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 2.0 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 1.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 1.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 2.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.8 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 1.6 | 6.4 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.6 | 1.9 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.5 | 2.6 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.4 | 6.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.4 | 2.6 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.3 | 5.8 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 0.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 1.5 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 0.9 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.4 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.7 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 1.0 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 1.8 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 1.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.6 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 3.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.4 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 0.6 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.6 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 0.8 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 1.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.3 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.4 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 1.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 1.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.5 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 1.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 3.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.5 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.3 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 1.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 2.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.3 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.3 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 1.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 2.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 2.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.2 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 1.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.5 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.9 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 8.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.2 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.3 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0070410 | JUN kinase binding(GO:0008432) co-SMAD binding(GO:0070410) |
| 0.0 | 0.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 5.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 1.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 2.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 3.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |