Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
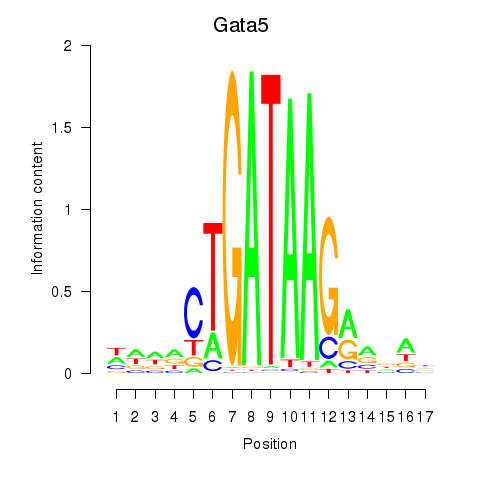
Results for Gata5
Z-value: 3.61
Transcription factors associated with Gata5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gata5
|
ENSMUSG00000015627.5 | GATA binding protein 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata5 | mm10_v2_chr2_-_180334665_180334704 | 0.89 | 4.0e-13 | Click! |
Activity profile of Gata5 motif
Sorted Z-values of Gata5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_84980458 | 39.10 |
ENSMUST00000028467.5
|
Prg2
|
proteoglycan 2, bone marrow |
| chr2_+_84988194 | 37.18 |
ENSMUST00000028466.5
|
Prg3
|
proteoglycan 3 |
| chr11_-_83286722 | 33.73 |
ENSMUST00000163961.2
|
Slfn14
|
schlafen family member 14 |
| chr17_+_40811089 | 32.04 |
ENSMUST00000024721.7
|
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr14_-_51057242 | 30.33 |
ENSMUST00000089798.2
|
Rnase12
|
ribonuclease, RNase A family, 12 (non-active) |
| chr1_+_40429563 | 29.69 |
ENSMUST00000174335.1
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr4_-_119189949 | 29.21 |
ENSMUST00000124626.1
|
Ermap
|
erythroblast membrane-associated protein |
| chr7_-_99238564 | 26.10 |
ENSMUST00000064231.7
|
Mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr7_-_103853199 | 24.34 |
ENSMUST00000033229.3
|
Hbb-y
|
hemoglobin Y, beta-like embryonic chain |
| chr1_-_132367879 | 23.53 |
ENSMUST00000142609.1
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr11_+_32276400 | 23.25 |
ENSMUST00000020531.2
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr11_+_32276893 | 22.27 |
ENSMUST00000145569.1
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr6_+_41354105 | 21.28 |
ENSMUST00000072103.5
|
Try10
|
trypsin 10 |
| chr17_-_28560704 | 21.09 |
ENSMUST00000114785.1
ENSMUST00000025062.3 |
Clps
|
colipase, pancreatic |
| chr8_+_84701430 | 20.73 |
ENSMUST00000037165.4
|
Lyl1
|
lymphoblastomic leukemia 1 |
| chr4_-_119190005 | 19.50 |
ENSMUST00000138395.1
ENSMUST00000156746.1 |
Ermap
|
erythroblast membrane-associated protein |
| chr9_+_21029373 | 18.95 |
ENSMUST00000001040.5
|
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
| chr17_-_26199008 | 18.80 |
ENSMUST00000142410.1
ENSMUST00000120333.1 ENSMUST00000039113.7 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr4_-_137430517 | 17.70 |
ENSMUST00000102522.4
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr13_+_108316332 | 17.36 |
ENSMUST00000051594.5
|
Depdc1b
|
DEP domain containing 1B |
| chr11_+_70639118 | 17.24 |
ENSMUST00000055184.6
ENSMUST00000108551.2 |
Gp1ba
|
glycoprotein 1b, alpha polypeptide |
| chr14_+_70457447 | 17.08 |
ENSMUST00000003561.3
|
Phyhip
|
phytanoyl-CoA hydroxylase interacting protein |
| chr8_-_85380964 | 16.68 |
ENSMUST00000122452.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr5_-_73191848 | 16.38 |
ENSMUST00000176910.1
|
Fryl
|
furry homolog-like (Drosophila) |
| chr6_-_41314700 | 16.01 |
ENSMUST00000064324.5
|
Try5
|
trypsin 5 |
| chr15_-_103251465 | 15.41 |
ENSMUST00000133600.1
ENSMUST00000134554.1 ENSMUST00000156927.1 ENSMUST00000149111.1 ENSMUST00000132836.1 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr11_+_58948890 | 14.31 |
ENSMUST00000078267.3
|
Hist3h2ba
|
histone cluster 3, H2ba |
| chr13_+_108316395 | 14.30 |
ENSMUST00000171178.1
|
Depdc1b
|
DEP domain containing 1B |
| chr11_-_102469839 | 13.50 |
ENSMUST00000103086.3
|
Itga2b
|
integrin alpha 2b |
| chr16_+_32186192 | 13.37 |
ENSMUST00000099990.3
|
Bex6
|
brain expressed gene 6 |
| chr7_-_103827922 | 13.34 |
ENSMUST00000023934.6
ENSMUST00000153218.1 |
Hbb-bs
|
hemoglobin, beta adult s chain |
| chr6_-_40585783 | 12.62 |
ENSMUST00000177178.1
ENSMUST00000129948.2 ENSMUST00000101491.4 |
Clec5a
|
C-type lectin domain family 5, member a |
| chr6_+_78370877 | 12.41 |
ENSMUST00000096904.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr11_+_95337012 | 12.30 |
ENSMUST00000037502.6
|
Fam117a
|
family with sequence similarity 117, member A |
| chr11_+_116531744 | 12.23 |
ENSMUST00000106387.2
ENSMUST00000100201.3 |
Sphk1
|
sphingosine kinase 1 |
| chr11_+_116532441 | 11.79 |
ENSMUST00000106386.1
ENSMUST00000145737.1 ENSMUST00000155102.1 ENSMUST00000063446.6 |
Sphk1
|
sphingosine kinase 1 |
| chr11_+_9191934 | 11.52 |
ENSMUST00000042740.6
|
Abca13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr17_+_34914459 | 10.89 |
ENSMUST00000007249.8
|
Slc44a4
|
solute carrier family 44, member 4 |
| chr4_-_137409777 | 10.56 |
ENSMUST00000024200.6
|
Gm13011
|
predicted gene 13011 |
| chr10_+_45577811 | 10.43 |
ENSMUST00000037044.6
|
Hace1
|
HECT domain and ankyrin repeat containing, E3 ubiquitin protein ligase 1 |
| chr6_+_41521782 | 10.28 |
ENSMUST00000070380.4
|
Prss2
|
protease, serine, 2 |
| chr7_-_127137807 | 10.26 |
ENSMUST00000049931.5
|
Spn
|
sialophorin |
| chr11_+_32296489 | 10.08 |
ENSMUST00000093207.3
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr3_+_105870858 | 9.87 |
ENSMUST00000164730.1
|
Adora3
|
adenosine A3 receptor |
| chr6_+_41302265 | 9.70 |
ENSMUST00000031913.4
|
Try4
|
trypsin 4 |
| chr3_-_14778452 | 9.12 |
ENSMUST00000094365.4
|
Car1
|
carbonic anhydrase 1 |
| chr7_-_121074501 | 9.09 |
ENSMUST00000047194.2
|
Igsf6
|
immunoglobulin superfamily, member 6 |
| chr13_-_62371936 | 8.54 |
ENSMUST00000107989.3
|
Gm3604
|
predicted gene 3604 |
| chr7_+_18718075 | 7.70 |
ENSMUST00000108481.1
ENSMUST00000051973.8 |
Psg22
|
pregnancy-specific glycoprotein 22 |
| chr3_-_14808358 | 7.56 |
ENSMUST00000181860.1
ENSMUST00000144327.2 |
Car1
|
carbonic anhydrase 1 |
| chr7_+_17087934 | 7.40 |
ENSMUST00000152671.1
|
Psg16
|
pregnancy specific glycoprotein 16 |
| chr14_+_61309753 | 6.84 |
ENSMUST00000055159.7
|
Arl11
|
ADP-ribosylation factor-like 11 |
| chr11_-_87359011 | 6.68 |
ENSMUST00000055438.4
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr2_-_73452666 | 6.66 |
ENSMUST00000151939.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr11_-_8950539 | 6.66 |
ENSMUST00000178195.1
|
Pkd1l1
|
polycystic kidney disease 1 like 1 |
| chr3_-_107221722 | 6.64 |
ENSMUST00000029504.8
|
Cym
|
chymosin |
| chr7_-_133702515 | 6.56 |
ENSMUST00000153698.1
|
Uros
|
uroporphyrinogen III synthase |
| chr13_-_62520451 | 6.27 |
ENSMUST00000082203.6
ENSMUST00000101547.4 |
Zfp934
|
zinc finger protein 934 |
| chr2_+_152427639 | 6.22 |
ENSMUST00000128737.1
|
6820408C15Rik
|
RIKEN cDNA 6820408C15 gene |
| chrX_-_8145713 | 6.12 |
ENSMUST00000115615.2
ENSMUST00000115616.1 ENSMUST00000115621.2 |
Rbm3
|
RNA binding motif protein 3 |
| chr2_+_72476159 | 6.06 |
ENSMUST00000102691.4
|
Cdca7
|
cell division cycle associated 7 |
| chr2_+_72476225 | 6.00 |
ENSMUST00000157019.1
|
Cdca7
|
cell division cycle associated 7 |
| chr4_+_103143052 | 5.92 |
ENSMUST00000106855.1
|
Mier1
|
mesoderm induction early response 1 homolog (Xenopus laevis |
| chr9_+_53771499 | 5.59 |
ENSMUST00000048670.8
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr7_-_30072801 | 5.59 |
ENSMUST00000183115.1
ENSMUST00000182919.1 ENSMUST00000183190.1 ENSMUST00000080834.8 |
Zfp82
|
zinc finger protein 82 |
| chr1_-_170927540 | 5.58 |
ENSMUST00000162136.1
ENSMUST00000162887.1 |
Fcrla
|
Fc receptor-like A |
| chr4_-_154928187 | 5.44 |
ENSMUST00000123514.1
|
Tnfrsf14
|
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
| chr17_+_17316078 | 5.43 |
ENSMUST00000105311.3
|
Gm6712
|
predicted gene 6712 |
| chr8_+_21734490 | 5.36 |
ENSMUST00000080533.5
|
Defa24
|
defensin, alpha, 24 |
| chr17_+_48346465 | 5.34 |
ENSMUST00000113237.3
|
Trem2
|
triggering receptor expressed on myeloid cells 2 |
| chr5_-_113830422 | 5.28 |
ENSMUST00000100874.4
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr9_+_124101944 | 5.27 |
ENSMUST00000171719.1
|
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr14_+_75455957 | 5.21 |
ENSMUST00000164848.1
|
Siah3
|
seven in absentia homolog 3 (Drosophila) |
| chr17_+_27342453 | 5.14 |
ENSMUST00000151398.1
|
Gm10505
|
predicted gene 10505 |
| chr2_+_4559742 | 5.03 |
ENSMUST00000176828.1
|
Frmd4a
|
FERM domain containing 4A |
| chr16_-_44558879 | 4.98 |
ENSMUST00000114634.1
|
Boc
|
biregional cell adhesion molecule-related/down-regulated by oncogenes (Cdon) binding protein |
| chr13_-_62777089 | 4.94 |
ENSMUST00000167516.2
|
Gm5141
|
predicted gene 5141 |
| chr12_-_27342696 | 4.93 |
ENSMUST00000079063.5
|
Sox11
|
SRY-box containing gene 11 |
| chr12_-_113307912 | 4.89 |
ENSMUST00000103418.1
|
Ighg2b
|
immunoglobulin heavy constant gamma 2B |
| chr7_+_126862431 | 4.76 |
ENSMUST00000132808.1
|
Hirip3
|
HIRA interacting protein 3 |
| chr12_+_83520436 | 4.69 |
ENSMUST00000021645.7
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chr3_+_105870898 | 4.68 |
ENSMUST00000010279.5
|
Adora3
|
adenosine A3 receptor |
| chr13_-_100317674 | 4.65 |
ENSMUST00000118574.1
|
Naip6
|
NLR family, apoptosis inhibitory protein 6 |
| chr11_-_101551837 | 4.43 |
ENSMUST00000017290.4
|
Brca1
|
breast cancer 1 |
| chr19_-_10678001 | 4.33 |
ENSMUST00000025647.5
|
Pga5
|
pepsinogen 5, group I |
| chr17_-_24527830 | 4.30 |
ENSMUST00000176353.1
ENSMUST00000176237.1 |
Traf7
|
TNF receptor-associated factor 7 |
| chr15_-_95528702 | 4.21 |
ENSMUST00000166170.1
|
Nell2
|
NEL-like 2 |
| chr2_+_32525013 | 4.20 |
ENSMUST00000150621.1
|
Gm13412
|
predicted gene 13412 |
| chr2_+_167062934 | 4.11 |
ENSMUST00000125674.1
|
1500012F01Rik
|
RIKEN cDNA 1500012F01 gene |
| chr1_+_135232045 | 3.95 |
ENSMUST00000110798.3
|
Gm4204
|
predicted gene 4204 |
| chr4_+_119637704 | 3.94 |
ENSMUST00000024015.2
|
Guca2a
|
guanylate cyclase activator 2a (guanylin) |
| chr10_+_79988584 | 3.72 |
ENSMUST00000004784.4
ENSMUST00000105374.1 |
Cnn2
|
calponin 2 |
| chr17_+_25369267 | 3.70 |
ENSMUST00000160377.1
ENSMUST00000160485.1 |
Tpsg1
|
tryptase gamma 1 |
| chr3_+_4211716 | 3.70 |
ENSMUST00000170943.1
|
Gm8775
|
predicted gene 8775 |
| chr1_-_136346074 | 3.54 |
ENSMUST00000048309.6
|
Camsap2
|
calmodulin regulated spectrin-associated protein family, member 2 |
| chr4_+_6191093 | 3.42 |
ENSMUST00000029907.5
|
Ubxn2b
|
UBX domain protein 2B |
| chr15_-_86186136 | 3.41 |
ENSMUST00000044332.9
|
Cerk
|
ceramide kinase |
| chr11_+_94936224 | 3.37 |
ENSMUST00000001547.7
|
Col1a1
|
collagen, type I, alpha 1 |
| chr6_+_34384218 | 3.36 |
ENSMUST00000038383.7
ENSMUST00000115051.1 |
Akr1b10
|
aldo-keto reductase family 1, member B10 (aldose reductase) |
| chr17_+_45523416 | 3.28 |
ENSMUST00000113547.1
|
Tcte1
|
t-complex-associated testis expressed 1 |
| chr6_+_48904979 | 3.27 |
ENSMUST00000162948.1
ENSMUST00000167529.1 |
Aoc1
|
amine oxidase, copper-containing 1 |
| chr2_+_150323702 | 3.26 |
ENSMUST00000133235.2
|
Gm10130
|
predicted gene 10130 |
| chr11_-_45955183 | 3.24 |
ENSMUST00000109254.1
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr11_+_4986824 | 2.95 |
ENSMUST00000009234.9
ENSMUST00000109897.1 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr7_+_101361997 | 2.92 |
ENSMUST00000133423.1
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chrX_-_139998519 | 2.86 |
ENSMUST00000113007.1
ENSMUST00000033810.7 ENSMUST00000113011.2 ENSMUST00000087400.5 |
Rbm41
|
RNA binding motif protein 41 |
| chr10_+_100488289 | 2.85 |
ENSMUST00000164751.1
|
Cep290
|
centrosomal protein 290 |
| chr1_+_85600672 | 2.79 |
ENSMUST00000080204.4
|
Sp140
|
Sp140 nuclear body protein |
| chr6_+_29279587 | 2.61 |
ENSMUST00000167131.1
|
Fam71f2
|
family with sequence similarity 71, member F2 |
| chr3_+_146121655 | 2.56 |
ENSMUST00000039450.4
|
Mcoln3
|
mucolipin 3 |
| chr11_-_118125944 | 2.49 |
ENSMUST00000124164.1
|
Dnah17
|
dynein, axonemal, heavy chain 17 |
| chr7_+_28833975 | 2.47 |
ENSMUST00000066723.8
|
Lgals4
|
lectin, galactose binding, soluble 4 |
| chr1_-_85270543 | 2.43 |
ENSMUST00000093506.5
ENSMUST00000064341.8 |
C130026I21Rik
|
RIKEN cDNA C130026I21 gene |
| chr12_-_69893162 | 2.34 |
ENSMUST00000049239.7
ENSMUST00000110570.1 |
Map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr1_+_51289106 | 2.28 |
ENSMUST00000051572.6
|
Sdpr
|
serum deprivation response |
| chr6_-_54566484 | 2.13 |
ENSMUST00000019268.4
|
Scrn1
|
secernin 1 |
| chr15_+_100154379 | 2.10 |
ENSMUST00000023768.6
ENSMUST00000108971.2 |
Dip2b
|
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
| chr15_+_6299797 | 2.10 |
ENSMUST00000159046.1
ENSMUST00000161040.1 |
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr9_-_41004599 | 1.98 |
ENSMUST00000180384.1
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr7_+_45621805 | 1.83 |
ENSMUST00000033100.4
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr8_-_12573311 | 1.78 |
ENSMUST00000180858.1
|
D630011A20Rik
|
RIKEN cDNA D630011A20 gene |
| chr7_-_126861648 | 1.71 |
ENSMUST00000129812.1
ENSMUST00000106342.1 |
Ino80e
|
INO80 complex subunit E |
| chr2_+_10370494 | 1.68 |
ENSMUST00000114862.1
ENSMUST00000114861.1 |
Sfmbt2
|
Scm-like with four mbt domains 2 |
| chr2_-_25224653 | 1.65 |
ENSMUST00000043584.4
|
Tubb4b
|
tubulin, beta 4B class IVB |
| chr16_+_36934976 | 1.64 |
ENSMUST00000023531.8
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chr3_-_93015669 | 1.60 |
ENSMUST00000107301.1
ENSMUST00000029521.4 |
Crct1
|
cysteine-rich C-terminal 1 |
| chr9_+_110798160 | 1.49 |
ENSMUST00000035715.6
|
Prss42
|
protease, serine, 42 |
| chr2_+_154656959 | 1.48 |
ENSMUST00000044277.9
|
Chmp4b
|
charged multivesicular body protein 4B |
| chr3_-_146839365 | 1.45 |
ENSMUST00000084614.3
|
Gm10288
|
predicted gene 10288 |
| chr5_+_87808082 | 1.41 |
ENSMUST00000072539.5
ENSMUST00000113279.1 ENSMUST00000101057.3 |
Csn1s2b
|
casein alpha s2-like B |
| chr7_-_126861828 | 1.40 |
ENSMUST00000106343.1
|
Ino80e
|
INO80 complex subunit E |
| chr6_+_81923645 | 1.39 |
ENSMUST00000043195.4
|
Gcfc2
|
GC-rich sequence DNA binding factor 2 |
| chr7_-_23947237 | 1.37 |
ENSMUST00000086013.2
|
Gm10175
|
predicted gene 10175 |
| chr13_-_100201961 | 1.30 |
ENSMUST00000167986.2
ENSMUST00000117913.1 |
Naip2
|
NLR family, apoptosis inhibitory protein 2 |
| chrX_+_166238901 | 1.28 |
ENSMUST00000112235.1
|
Gpm6b
|
glycoprotein m6b |
| chr7_-_14123042 | 1.28 |
ENSMUST00000098809.2
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr10_-_130127055 | 1.27 |
ENSMUST00000074161.1
|
Olfr824
|
olfactory receptor 824 |
| chr4_-_11981265 | 1.23 |
ENSMUST00000098260.2
|
Gm10604
|
predicted gene 10604 |
| chr11_+_70017199 | 1.21 |
ENSMUST00000133140.1
|
Dlg4
|
discs, large homolog 4 (Drosophila) |
| chr15_+_39745926 | 1.20 |
ENSMUST00000022913.4
|
Dcstamp
|
dentrocyte expressed seven transmembrane protein |
| chr11_-_109995743 | 1.20 |
ENSMUST00000106669.2
|
Abca8b
|
ATP-binding cassette, sub-family A (ABC1), member 8b |
| chrX_-_37110257 | 1.17 |
ENSMUST00000076265.6
|
Upf3b
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr9_+_38877126 | 1.10 |
ENSMUST00000078289.2
|
Olfr926
|
olfactory receptor 926 |
| chr6_+_135011609 | 1.10 |
ENSMUST00000032326.4
ENSMUST00000130851.1 ENSMUST00000154558.1 |
Ddx47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr8_+_19682268 | 1.04 |
ENSMUST00000153710.1
ENSMUST00000127799.1 |
Gm6483
|
predicted gene 6483 |
| chr7_-_135528645 | 1.03 |
ENSMUST00000053716.7
|
Clrn3
|
clarin 3 |
| chr12_+_69241832 | 1.02 |
ENSMUST00000063445.6
|
Klhdc1
|
kelch domain containing 1 |
| chr9_-_58201705 | 1.01 |
ENSMUST00000163200.1
ENSMUST00000165276.1 |
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr11_-_109995775 | 1.00 |
ENSMUST00000020948.8
|
Abca8b
|
ATP-binding cassette, sub-family A (ABC1), member 8b |
| chr1_+_71652837 | 1.00 |
ENSMUST00000097699.2
|
Apol7d
|
apolipoprotein L 7d |
| chr6_-_34317442 | 0.99 |
ENSMUST00000154655.1
ENSMUST00000102980.4 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr2_+_164698501 | 0.99 |
ENSMUST00000017454.7
|
Spint4
|
serine protease inhibitor, Kunitz type 4 |
| chr11_+_90030295 | 0.97 |
ENSMUST00000092788.3
|
Tmem100
|
transmembrane protein 100 |
| chr8_+_20136455 | 0.94 |
ENSMUST00000179299.1
ENSMUST00000096485.4 |
Gm21811
|
predicted gene, 21811 |
| chr2_+_71786923 | 0.91 |
ENSMUST00000112101.1
ENSMUST00000028522.3 |
Itga6
|
integrin alpha 6 |
| chr7_-_18532225 | 0.89 |
ENSMUST00000094795.4
|
Psg25
|
pregnancy-specific glycoprotein 25 |
| chr12_+_95692212 | 0.88 |
ENSMUST00000057324.3
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr3_+_95427575 | 0.86 |
ENSMUST00000181809.1
|
Gm4349
|
predicted gene 4349 |
| chr13_+_3837757 | 0.86 |
ENSMUST00000042219.4
|
Calm4
|
calmodulin 4 |
| chr3_-_95106907 | 0.84 |
ENSMUST00000107233.2
|
Pip5k1a
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 alpha |
| chr16_+_92612755 | 0.82 |
ENSMUST00000180989.1
|
Gm26626
|
predicted gene, 26626 |
| chr15_-_77153772 | 0.82 |
ENSMUST00000166610.1
ENSMUST00000111581.2 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chrX_-_134111852 | 0.80 |
ENSMUST00000033610.6
|
Nox1
|
NADPH oxidase 1 |
| chr16_+_57549232 | 0.79 |
ENSMUST00000159414.1
|
Filip1l
|
filamin A interacting protein 1-like |
| chr19_-_38819156 | 0.79 |
ENSMUST00000025963.7
|
Noc3l
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr18_+_7869707 | 0.78 |
ENSMUST00000166062.1
ENSMUST00000169010.1 |
Wac
|
WW domain containing adaptor with coiled-coil |
| chrX_-_134111708 | 0.78 |
ENSMUST00000159259.1
ENSMUST00000113275.3 |
Nox1
|
NADPH oxidase 1 |
| chrX_-_37104523 | 0.76 |
ENSMUST00000130324.1
|
Upf3b
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr4_-_106727930 | 0.76 |
ENSMUST00000106770.1
ENSMUST00000145044.1 |
Mroh7
|
maestro heat-like repeat family member 7 |
| chrX_+_112495266 | 0.73 |
ENSMUST00000026602.2
ENSMUST00000113412.2 |
2010106E10Rik
|
RIKEN cDNA 2010106E10 gene |
| chr2_-_7081207 | 0.71 |
ENSMUST00000114923.2
ENSMUST00000182706.1 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr2_-_7081256 | 0.70 |
ENSMUST00000183209.1
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr12_-_103425780 | 0.67 |
ENSMUST00000110001.2
ENSMUST00000044923.7 |
Ddx24
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 24 |
| chr17_+_74489492 | 0.63 |
ENSMUST00000024873.6
|
Yipf4
|
Yip1 domain family, member 4 |
| chr14_-_101200069 | 0.53 |
ENSMUST00000057718.4
|
1700110M21Rik
|
RIKEN cDNA 1700110M21 gene |
| chr14_-_52036143 | 0.45 |
ENSMUST00000052560.4
|
Olfr221
|
olfactory receptor 221 |
| chrX_+_52988119 | 0.41 |
ENSMUST00000026723.8
|
Hprt
|
hypoxanthine guanine phosphoribosyl transferase |
| chr7_-_46672537 | 0.31 |
ENSMUST00000049298.7
|
Tph1
|
tryptophan hydroxylase 1 |
| chr4_-_59438633 | 0.31 |
ENSMUST00000040166.7
ENSMUST00000107544.1 |
Susd1
|
sushi domain containing 1 |
| chr11_+_87853207 | 0.30 |
ENSMUST00000038196.6
|
Mks1
|
Meckel syndrome, type 1 |
| chr6_-_147243794 | 0.30 |
ENSMUST00000153786.1
|
Gm15767
|
predicted gene 15767 |
| chr12_-_112674193 | 0.28 |
ENSMUST00000001780.3
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr7_+_104003259 | 0.20 |
ENSMUST00000098184.1
|
Olfr638
|
olfactory receptor 638 |
| chr19_-_29523159 | 0.19 |
ENSMUST00000180986.1
|
A930007I19Rik
|
RIKEN cDNA A930007I19 gene |
| chr1_+_172376528 | 0.18 |
ENSMUST00000052455.2
|
Pigm
|
phosphatidylinositol glycan anchor biosynthesis, class M |
| chr15_+_73834561 | 0.12 |
ENSMUST00000154520.1
|
Gm6569
|
predicted gene 6569 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gata5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.0 | 39.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 10.7 | 32.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 9.3 | 37.2 | GO:0045575 | basophil activation(GO:0045575) |
| 8.7 | 69.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 8.7 | 26.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 4.0 | 24.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 3.6 | 10.9 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 3.4 | 10.3 | GO:0002865 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) negative regulation of hypersensitivity(GO:0002884) |
| 3.0 | 20.7 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 2.7 | 27.2 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 2.3 | 13.6 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 2.2 | 6.7 | GO:0003127 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 1.9 | 7.7 | GO:2001201 | regulation of transforming growth factor-beta secretion(GO:2001201) |
| 1.9 | 33.7 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 1.8 | 5.3 | GO:0002582 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 1.8 | 5.3 | GO:1902567 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) astrocyte chemotaxis(GO:0035700) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) regulation of astrocyte chemotaxis(GO:2000458) |
| 1.6 | 6.6 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 1.6 | 16.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 1.5 | 4.4 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 1.5 | 14.6 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 1.3 | 15.4 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 1.2 | 4.9 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 1.0 | 5.2 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 1.0 | 1.0 | GO:0051595 | response to methylglyoxal(GO:0051595) |
| 1.0 | 4.9 | GO:0016068 | regulation of type I hypersensitivity(GO:0001810) positive regulation of type I hypersensitivity(GO:0001812) type I hypersensitivity(GO:0016068) |
| 0.7 | 17.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.7 | 2.0 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.6 | 10.3 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.6 | 12.6 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.6 | 5.5 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.6 | 20.4 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.6 | 3.4 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.5 | 5.4 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.5 | 21.4 | GO:0032094 | response to food(GO:0032094) |
| 0.5 | 5.9 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.4 | 6.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.4 | 6.0 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.4 | 4.9 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.4 | 16.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.4 | 4.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.3 | 2.1 | GO:0035026 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) leading edge cell differentiation(GO:0035026) |
| 0.3 | 4.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 20.5 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.3 | 1.2 | GO:0034241 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.3 | 36.1 | GO:0007586 | digestion(GO:0007586) |
| 0.3 | 3.3 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.2 | 10.1 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.2 | 6.7 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.2 | 1.3 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 31.7 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.2 | 0.8 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 3.7 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.2 | 0.9 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.2 | 5.4 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.2 | 13.5 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.2 | 1.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.2 | 1.0 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.2 | 0.3 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 6.1 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 10.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 2.7 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 0.4 | GO:0046098 | purine nucleobase salvage(GO:0043096) guanine metabolic process(GO:0046098) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 | 3.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 5.0 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 1.6 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.1 | 14.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 5.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 1.2 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 0.9 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 2.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 6.2 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 1.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 3.2 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 2.5 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.3 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 1.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.8 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.8 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 3.6 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 2.1 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 4.4 | GO:0008380 | RNA splicing(GO:0008380) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.7 | 93.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 1.2 | 15.5 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 1.1 | 3.4 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.9 | 6.0 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.7 | 17.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.7 | 4.4 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.7 | 3.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.5 | 2.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.4 | 12.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.3 | 1.4 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.3 | 14.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.3 | 1.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.3 | 1.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.3 | 4.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 3.1 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.2 | 4.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 3.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 14.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 2.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 1.6 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 1.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 16.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 14.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 25.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 6.7 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 5.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 29.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 3.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 1.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 5.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 16.4 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 6.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 23.0 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 6.1 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 1.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 46.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 7.9 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 7.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 34.3 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 12.6 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 10.6 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.9 | 29.7 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 9.4 | 37.7 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 7.6 | 45.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 5.2 | 26.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 5.0 | 10.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 3.4 | 24.0 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 2.7 | 13.5 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 2.4 | 16.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 2.1 | 16.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.8 | 5.3 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 1.7 | 18.8 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 1.7 | 32.0 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 1.6 | 10.9 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 1.3 | 14.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 1.1 | 3.4 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 1.1 | 3.4 | GO:0070401 | NADP+ binding(GO:0070401) |
| 1.1 | 6.6 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 1.1 | 3.2 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 1.0 | 7.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.8 | 3.9 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.7 | 6.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.6 | 5.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.5 | 12.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.5 | 32.2 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.5 | 30.3 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.4 | 3.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 11.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.4 | 3.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.4 | 2.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.3 | 70.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.3 | 15.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 1.6 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.3 | 84.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.3 | 2.9 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.2 | 3.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 4.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 1.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 2.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.2 | 1.3 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.2 | 10.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 22.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 2.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 5.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 1.2 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 1.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 10.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 5.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 6.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.8 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 6.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 2.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 28.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 0.3 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 4.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 4.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 14.9 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.1 | 4.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 6.7 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 8.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 6.8 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 4.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 3.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.8 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 8.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 2.6 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 76.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 1.1 | 36.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.9 | 24.0 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.7 | 10.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.6 | 14.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.3 | 26.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 5.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 3.4 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 8.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 3.4 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 5.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 12.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 24.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 14.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.8 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 2.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 2.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 15.6 | REACTOME DEFENSINS | Genes involved in Defensins |
| 1.4 | 20.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 1.3 | 42.9 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.8 | 13.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.7 | 14.6 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.5 | 24.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.5 | 21.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.4 | 20.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 10.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.3 | 2.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 4.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 3.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 5.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 4.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 30.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 5.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 5.7 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 1.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 2.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |