Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
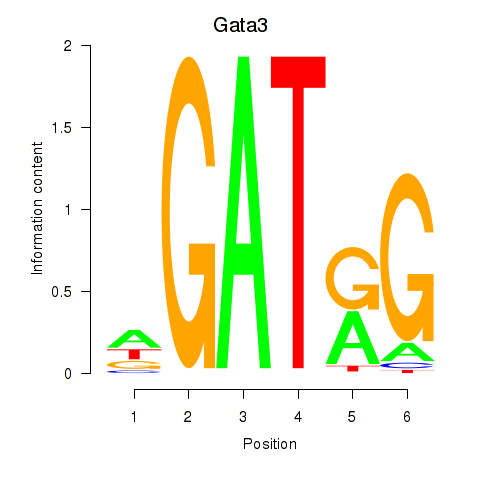
Results for Gata3
Z-value: 1.82
Transcription factors associated with Gata3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gata3
|
ENSMUSG00000015619.10 | GATA binding protein 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata3 | mm10_v2_chr2_-_9878580_9878607 | -0.05 | 7.6e-01 | Click! |
Activity profile of Gata3 motif
Sorted Z-values of Gata3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_32276400 | 5.86 |
ENSMUST00000020531.2
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr11_+_32276893 | 4.75 |
ENSMUST00000145569.1
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr7_-_103843154 | 3.24 |
ENSMUST00000063957.4
|
Hbb-bh1
|
hemoglobin Z, beta-like embryonic chain |
| chr13_-_76056996 | 3.24 |
ENSMUST00000056130.4
|
Gpr150
|
G protein-coupled receptor 150 |
| chr7_-_103853199 | 3.20 |
ENSMUST00000033229.3
|
Hbb-y
|
hemoglobin Y, beta-like embryonic chain |
| chr7_+_103937382 | 3.19 |
ENSMUST00000098189.1
|
Olfr632
|
olfactory receptor 632 |
| chr13_+_23746734 | 2.52 |
ENSMUST00000099703.2
|
Hist1h2bb
|
histone cluster 1, H2bb |
| chr4_+_13751297 | 2.43 |
ENSMUST00000105566.2
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chrX_+_101449078 | 2.39 |
ENSMUST00000033674.5
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr15_-_103251465 | 2.33 |
ENSMUST00000133600.1
ENSMUST00000134554.1 ENSMUST00000156927.1 ENSMUST00000149111.1 ENSMUST00000132836.1 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr7_-_30534180 | 2.21 |
ENSMUST00000044338.4
|
Arhgap33
|
Rho GTPase activating protein 33 |
| chr15_-_103252810 | 2.14 |
ENSMUST00000154510.1
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr7_-_25369712 | 2.11 |
ENSMUST00000167591.1
ENSMUST00000076276.3 |
Cnfn
|
cornifelin |
| chr14_+_31208309 | 2.10 |
ENSMUST00000169169.1
|
Tnnc1
|
troponin C, cardiac/slow skeletal |
| chr15_-_101924725 | 2.08 |
ENSMUST00000023797.6
|
Krt4
|
keratin 4 |
| chr13_+_21722057 | 2.00 |
ENSMUST00000110476.3
|
Hist1h2bm
|
histone cluster 1, H2bm |
| chr18_+_61687911 | 2.00 |
ENSMUST00000025471.2
|
Il17b
|
interleukin 17B |
| chr11_+_87760533 | 1.97 |
ENSMUST00000039627.5
ENSMUST00000100644.3 |
Bzrap1
|
benzodiazepine receptor associated protein 1 |
| chr14_-_70627008 | 1.94 |
ENSMUST00000110984.2
|
Dmtn
|
dematin actin binding protein |
| chr15_-_101694299 | 1.90 |
ENSMUST00000023788.6
|
Krt6a
|
keratin 6A |
| chr3_+_103832562 | 1.86 |
ENSMUST00000062945.5
|
Bcl2l15
|
BCLl2-like 15 |
| chr11_+_95824469 | 1.82 |
ENSMUST00000150134.1
ENSMUST00000054173.3 |
Phospho1
|
phosphatase, orphan 1 |
| chr2_+_129228022 | 1.74 |
ENSMUST00000148548.1
|
A730036I17Rik
|
RIKEN cDNA A730036I17 gene |
| chr2_+_25372315 | 1.73 |
ENSMUST00000028329.6
ENSMUST00000114293.2 ENSMUST00000100323.2 |
Sapcd2
|
suppressor APC domain containing 2 |
| chr11_-_100121558 | 1.73 |
ENSMUST00000007275.2
|
Krt13
|
keratin 13 |
| chr14_-_37110087 | 1.65 |
ENSMUST00000179488.1
|
2610528A11Rik
|
RIKEN cDNA 2610528A11 gene |
| chr7_+_30787897 | 1.63 |
ENSMUST00000098559.1
|
Krtdap
|
keratinocyte differentiation associated protein |
| chr5_+_115908644 | 1.62 |
ENSMUST00000141101.1
|
Cit
|
citron |
| chr13_-_3804307 | 1.60 |
ENSMUST00000077698.3
|
Calml3
|
calmodulin-like 3 |
| chr13_-_21753851 | 1.53 |
ENSMUST00000074752.2
|
Hist1h2ak
|
histone cluster 1, H2ak |
| chr10_-_129902726 | 1.52 |
ENSMUST00000071557.1
|
Olfr815
|
olfactory receptor 815 |
| chr11_+_5569679 | 1.52 |
ENSMUST00000109856.1
ENSMUST00000109855.1 ENSMUST00000118112.2 |
Ankrd36
|
ankyrin repeat domain 36 |
| chr11_+_101316917 | 1.49 |
ENSMUST00000151385.1
|
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr2_+_155517948 | 1.47 |
ENSMUST00000029135.8
ENSMUST00000065973.2 ENSMUST00000103142.5 |
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr4_+_117849193 | 1.46 |
ENSMUST00000132043.2
ENSMUST00000169990.1 |
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr4_+_106733883 | 1.45 |
ENSMUST00000047620.2
|
Fam151a
|
family with sequence simliarity 151, member A |
| chrX_-_104671048 | 1.45 |
ENSMUST00000042070.5
|
Zdhhc15
|
zinc finger, DHHC domain containing 15 |
| chr10_+_43579161 | 1.44 |
ENSMUST00000058714.8
|
Cd24a
|
CD24a antigen |
| chr10_+_14523062 | 1.41 |
ENSMUST00000096020.5
|
Gm10335
|
predicted gene 10335 |
| chr1_-_132367879 | 1.40 |
ENSMUST00000142609.1
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr2_-_121036750 | 1.36 |
ENSMUST00000023987.5
|
Epb4.2
|
erythrocyte protein band 4.2 |
| chr9_-_72491939 | 1.36 |
ENSMUST00000185151.1
ENSMUST00000085358.5 ENSMUST00000184125.1 ENSMUST00000183574.1 ENSMUST00000184831.1 |
Tex9
|
testis expressed gene 9 |
| chr9_-_44342332 | 1.35 |
ENSMUST00000097558.3
|
Hmbs
|
hydroxymethylbilane synthase |
| chr2_+_91035613 | 1.35 |
ENSMUST00000111445.3
ENSMUST00000111446.3 ENSMUST00000050323.5 |
Rapsn
|
receptor-associated protein of the synapse |
| chr8_+_105297663 | 1.35 |
ENSMUST00000015003.8
|
E2f4
|
E2F transcription factor 4 |
| chr16_-_4523056 | 1.32 |
ENSMUST00000090500.3
ENSMUST00000023161.7 |
Srl
|
sarcalumenin |
| chr12_+_76837408 | 1.32 |
ENSMUST00000041008.9
|
Fntb
|
farnesyltransferase, CAAX box, beta |
| chr4_+_115059507 | 1.32 |
ENSMUST00000162489.1
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr17_+_28207778 | 1.30 |
ENSMUST00000002327.5
|
Def6
|
differentially expressed in FDCP 6 |
| chr12_-_10900296 | 1.30 |
ENSMUST00000085735.2
|
Pgk1-rs7
|
phosphoglycerate kinase-1, related sequence-7 |
| chr7_-_103827922 | 1.29 |
ENSMUST00000023934.6
ENSMUST00000153218.1 |
Hbb-bs
|
hemoglobin, beta adult s chain |
| chrX_-_141874870 | 1.27 |
ENSMUST00000182079.1
|
Gm15294
|
predicted gene 15294 |
| chr11_+_101325063 | 1.27 |
ENSMUST00000041095.7
ENSMUST00000107264.1 |
Aoc2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr7_-_102477902 | 1.26 |
ENSMUST00000061482.5
|
Olfr543
|
olfactory receptor 543 |
| chr3_+_96269695 | 1.26 |
ENSMUST00000051089.3
ENSMUST00000177113.1 |
Hist2h2bb
|
histone cluster 2, H2bb |
| chr8_+_84970068 | 1.26 |
ENSMUST00000164807.1
|
Prdx2
|
peroxiredoxin 2 |
| chr8_+_21378560 | 1.25 |
ENSMUST00000170275.2
|
Defa2
|
defensin, alpha, 2 |
| chr8_-_121907678 | 1.24 |
ENSMUST00000045557.9
|
Slc7a5
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 |
| chr6_-_124464772 | 1.24 |
ENSMUST00000008297.4
|
Clstn3
|
calsyntenin 3 |
| chr8_+_84969824 | 1.22 |
ENSMUST00000125893.1
|
Prdx2
|
peroxiredoxin 2 |
| chr2_+_163054682 | 1.21 |
ENSMUST00000018005.3
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr4_-_87806296 | 1.21 |
ENSMUST00000126353.1
ENSMUST00000149357.1 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr19_+_3992752 | 1.19 |
ENSMUST00000041871.7
|
Tbx10
|
T-box 10 |
| chr2_+_84734050 | 1.19 |
ENSMUST00000090729.2
|
Ypel4
|
yippee-like 4 (Drosophila) |
| chr4_-_132533488 | 1.19 |
ENSMUST00000152993.1
ENSMUST00000067496.6 |
Atpif1
|
ATPase inhibitory factor 1 |
| chr2_-_25224653 | 1.19 |
ENSMUST00000043584.4
|
Tubb4b
|
tubulin, beta 4B class IVB |
| chr13_-_21833575 | 1.19 |
ENSMUST00000081342.5
|
Hist1h2ap
|
histone cluster 1, H2ap |
| chr11_-_69605829 | 1.18 |
ENSMUST00000047889.6
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr7_-_133709051 | 1.18 |
ENSMUST00000124759.1
ENSMUST00000106144.1 |
Uros
|
uroporphyrinogen III synthase |
| chr11_-_84828953 | 1.18 |
ENSMUST00000047560.7
|
Dhrs11
|
dehydrogenase/reductase (SDR family) member 11 |
| chr7_+_103928825 | 1.17 |
ENSMUST00000106863.1
|
Olfr631
|
olfactory receptor 631 |
| chr4_-_43046196 | 1.17 |
ENSMUST00000036462.5
|
Fam214b
|
family with sequence similarity 214, member B |
| chr3_-_108085346 | 1.17 |
ENSMUST00000078912.5
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr13_+_91461050 | 1.16 |
ENSMUST00000004094.8
ENSMUST00000042122.8 |
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr4_-_117178726 | 1.15 |
ENSMUST00000153953.1
ENSMUST00000106436.1 |
Kif2c
|
kinesin family member 2C |
| chr15_+_74516196 | 1.15 |
ENSMUST00000042035.9
|
Bai1
|
brain-specific angiogenesis inhibitor 1 |
| chr4_-_87806276 | 1.14 |
ENSMUST00000148059.1
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr8_+_84969767 | 1.13 |
ENSMUST00000109733.1
|
Prdx2
|
peroxiredoxin 2 |
| chr7_-_133709069 | 1.12 |
ENSMUST00000106145.3
|
Uros
|
uroporphyrinogen III synthase |
| chr9_-_107985863 | 1.12 |
ENSMUST00000048568.4
|
Fam212a
|
family with sequence similarity 212, member A |
| chr19_+_10015016 | 1.12 |
ENSMUST00000137637.1
ENSMUST00000149967.1 |
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chrX_+_56454871 | 1.12 |
ENSMUST00000039374.2
ENSMUST00000101553.2 |
Ddx26b
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr11_+_58948890 | 1.12 |
ENSMUST00000078267.3
|
Hist3h2ba
|
histone cluster 3, H2ba |
| chr19_+_9283231 | 1.12 |
ENSMUST00000088040.4
|
Pcna-ps2
|
proliferating cell nuclear antigen pseudogene 2 |
| chr13_-_97747399 | 1.11 |
ENSMUST00000144993.1
|
5330416C01Rik
|
RIKEN cDNA 5330416C01 gene |
| chr7_-_133708958 | 1.11 |
ENSMUST00000106146.1
|
Uros
|
uroporphyrinogen III synthase |
| chr3_+_114030532 | 1.10 |
ENSMUST00000123619.1
ENSMUST00000092155.5 |
Col11a1
|
collagen, type XI, alpha 1 |
| chr2_-_121037048 | 1.09 |
ENSMUST00000102490.3
|
Epb4.2
|
erythrocyte protein band 4.2 |
| chr2_-_164356067 | 1.07 |
ENSMUST00000165980.1
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr1_-_66935333 | 1.07 |
ENSMUST00000120415.1
ENSMUST00000119429.1 |
Myl1
|
myosin, light polypeptide 1 |
| chr7_-_115846080 | 1.07 |
ENSMUST00000166207.1
|
Sox6
|
SRY-box containing gene 6 |
| chr2_-_84822546 | 1.07 |
ENSMUST00000028471.5
|
Smtnl1
|
smoothelin-like 1 |
| chr12_-_4841583 | 1.07 |
ENSMUST00000020964.5
|
Fkbp1b
|
FK506 binding protein 1b |
| chr11_-_100527862 | 1.06 |
ENSMUST00000107385.1
|
Acly
|
ATP citrate lyase |
| chr11_-_109828021 | 1.06 |
ENSMUST00000020941.4
|
1700012B07Rik
|
RIKEN cDNA 1700012B07 gene |
| chr3_+_19644452 | 1.05 |
ENSMUST00000029139.7
|
Trim55
|
tripartite motif-containing 55 |
| chr11_-_102897123 | 1.05 |
ENSMUST00000067444.3
|
Gfap
|
glial fibrillary acidic protein |
| chr11_+_62248977 | 1.05 |
ENSMUST00000018644.2
|
Adora2b
|
adenosine A2b receptor |
| chr6_+_120093348 | 1.03 |
ENSMUST00000112711.2
|
Ninj2
|
ninjurin 2 |
| chr17_+_28530834 | 1.03 |
ENSMUST00000025060.2
|
Armc12
|
armadillo repeat containing 12 |
| chr11_-_5803733 | 1.02 |
ENSMUST00000020768.3
|
Pgam2
|
phosphoglycerate mutase 2 |
| chr11_-_102897146 | 1.01 |
ENSMUST00000077902.4
|
Gfap
|
glial fibrillary acidic protein |
| chr1_-_160153571 | 1.01 |
ENSMUST00000039178.5
|
Tnn
|
tenascin N |
| chr9_+_72438534 | 1.01 |
ENSMUST00000034746.8
|
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr7_-_116038734 | 1.00 |
ENSMUST00000166877.1
|
Sox6
|
SRY-box containing gene 6 |
| chr10_+_115817247 | 0.99 |
ENSMUST00000035563.7
ENSMUST00000080630.3 ENSMUST00000179196.1 |
Tspan8
|
tetraspanin 8 |
| chr1_+_86021935 | 0.99 |
ENSMUST00000052854.6
ENSMUST00000125083.1 ENSMUST00000152501.1 ENSMUST00000113344.1 ENSMUST00000130504.1 ENSMUST00000153247.2 |
Spata3
|
spermatogenesis associated 3 |
| chr14_-_79662084 | 0.99 |
ENSMUST00000165835.1
|
Lect1
|
leukocyte cell derived chemotaxin 1 |
| chr9_+_120114850 | 0.98 |
ENSMUST00000135514.1
|
Slc25a38
|
solute carrier family 25, member 38 |
| chr2_+_84839395 | 0.97 |
ENSMUST00000146816.1
ENSMUST00000028469.7 |
Slc43a1
|
solute carrier family 43, member 1 |
| chr7_-_98145472 | 0.97 |
ENSMUST00000098281.2
|
Omp
|
olfactory marker protein |
| chr2_+_111780313 | 0.96 |
ENSMUST00000054004.1
|
Olfr1302
|
olfactory receptor 1302 |
| chr17_+_85028347 | 0.96 |
ENSMUST00000024944.7
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr5_-_139736291 | 0.96 |
ENSMUST00000044642.10
|
Micall2
|
MICAL-like 2 |
| chr18_-_60501983 | 0.95 |
ENSMUST00000042710.6
|
Smim3
|
small integral membrane protein 3 |
| chr2_-_150668198 | 0.94 |
ENSMUST00000028944.3
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr9_-_108083330 | 0.94 |
ENSMUST00000159372.1
ENSMUST00000160249.1 |
Rnf123
|
ring finger protein 123 |
| chr7_-_116031047 | 0.94 |
ENSMUST00000106612.1
|
Sox6
|
SRY-box containing gene 6 |
| chr3_+_96268654 | 0.93 |
ENSMUST00000098843.2
|
Hist2h3b
|
histone cluster 2, H3b |
| chr17_+_37529957 | 0.92 |
ENSMUST00000097325.3
|
Olfr111
|
olfactory receptor 111 |
| chr8_+_123102344 | 0.92 |
ENSMUST00000000756.5
|
Rpl13
|
ribosomal protein L13 |
| chr3_-_116253467 | 0.91 |
ENSMUST00000090473.5
|
Gpr88
|
G-protein coupled receptor 88 |
| chr11_-_74724670 | 0.91 |
ENSMUST00000021091.8
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr4_-_117929466 | 0.91 |
ENSMUST00000097913.2
|
Artn
|
artemin |
| chr17_+_35821675 | 0.90 |
ENSMUST00000003635.6
|
Ier3
|
immediate early response 3 |
| chr13_-_97747373 | 0.90 |
ENSMUST00000123535.1
|
5330416C01Rik
|
RIKEN cDNA 5330416C01 gene |
| chr7_+_45514619 | 0.89 |
ENSMUST00000107761.1
|
Tulp2
|
tubby-like protein 2 |
| chr13_+_23535411 | 0.88 |
ENSMUST00000080859.5
|
Hist1h3g
|
histone cluster 1, H3g |
| chr14_-_31577318 | 0.88 |
ENSMUST00000112027.2
|
Colq
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr2_+_103970115 | 0.88 |
ENSMUST00000111143.1
ENSMUST00000138815.1 |
Lmo2
|
LIM domain only 2 |
| chr17_-_35066170 | 0.87 |
ENSMUST00000174190.1
ENSMUST00000097337.1 |
AU023871
|
expressed sequence AU023871 |
| chr7_-_115824699 | 0.87 |
ENSMUST00000169129.1
|
Sox6
|
SRY-box containing gene 6 |
| chr7_+_104003259 | 0.87 |
ENSMUST00000098184.1
|
Olfr638
|
olfactory receptor 638 |
| chr11_-_95842409 | 0.87 |
ENSMUST00000059026.3
|
Abi3
|
ABI gene family, member 3 |
| chr7_+_25152456 | 0.86 |
ENSMUST00000098678.1
|
D930028M14Rik
|
RIKEN cDNA D930028M14 gene |
| chr4_-_156200818 | 0.86 |
ENSMUST00000085425.4
|
Isg15
|
ISG15 ubiquitin-like modifier |
| chr4_-_43045686 | 0.86 |
ENSMUST00000107956.1
ENSMUST00000107957.1 |
Fam214b
|
family with sequence similarity 214, member B |
| chr11_-_82829024 | 0.85 |
ENSMUST00000021036.6
ENSMUST00000074515.4 ENSMUST00000103218.2 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr7_+_78914216 | 0.85 |
ENSMUST00000120331.2
|
Isg20
|
interferon-stimulated protein |
| chr1_-_52232296 | 0.85 |
ENSMUST00000114512.1
|
Gls
|
glutaminase |
| chr4_-_63172118 | 0.85 |
ENSMUST00000030042.2
|
Kif12
|
kinesin family member 12 |
| chr19_-_11623643 | 0.85 |
ENSMUST00000164792.1
ENSMUST00000025583.5 |
Ms4a2
|
membrane-spanning 4-domains, subfamily A, member 2 |
| chr16_-_76022266 | 0.85 |
ENSMUST00000114240.1
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr16_+_13986596 | 0.84 |
ENSMUST00000056521.5
ENSMUST00000118412.1 ENSMUST00000131608.1 |
2900011O08Rik
|
RIKEN cDNA 2900011O08 gene |
| chr14_+_55491062 | 0.84 |
ENSMUST00000076236.5
|
Lrrc16b
|
leucine rich repeat containing 16B |
| chr15_-_95528702 | 0.84 |
ENSMUST00000166170.1
|
Nell2
|
NEL-like 2 |
| chr1_-_16770138 | 0.84 |
ENSMUST00000071842.8
|
Gm5828
|
predicted gene 5828 |
| chr6_-_122609964 | 0.83 |
ENSMUST00000032211.4
|
Gdf3
|
growth differentiation factor 3 |
| chr2_+_103957976 | 0.82 |
ENSMUST00000156813.1
ENSMUST00000170926.1 |
Lmo2
|
LIM domain only 2 |
| chr4_+_127172866 | 0.82 |
ENSMUST00000106094.2
|
Dlgap3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr13_-_23571151 | 0.81 |
ENSMUST00000102969.3
|
Hist1h2ae
|
histone cluster 1, H2ae |
| chr11_-_6230127 | 0.81 |
ENSMUST00000004505.2
|
Npc1l1
|
NPC1-like 1 |
| chr6_-_34979054 | 0.81 |
ENSMUST00000144079.1
|
2010107G12Rik
|
RIKEN cDNA 2010107G12 gene |
| chr17_+_29490812 | 0.80 |
ENSMUST00000024811.6
|
Pim1
|
proviral integration site 1 |
| chr11_+_53433299 | 0.79 |
ENSMUST00000018382.6
|
Gdf9
|
growth differentiation factor 9 |
| chr11_+_102604370 | 0.79 |
ENSMUST00000057893.5
|
Fzd2
|
frizzled homolog 2 (Drosophila) |
| chr18_+_61953048 | 0.79 |
ENSMUST00000051720.5
|
Sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr4_-_117182623 | 0.79 |
ENSMUST00000065896.2
|
Kif2c
|
kinesin family member 2C |
| chr2_-_126491566 | 0.79 |
ENSMUST00000040149.6
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr6_-_117214048 | 0.79 |
ENSMUST00000170447.1
|
Rpl28-ps4
|
ribosomal protein L28, pseudogene 4 |
| chr11_+_4236411 | 0.78 |
ENSMUST00000075221.2
|
Osm
|
oncostatin M |
| chr9_+_122951051 | 0.78 |
ENSMUST00000040717.5
|
Kif15
|
kinesin family member 15 |
| chr6_+_41684414 | 0.78 |
ENSMUST00000031900.5
|
1700034O15Rik
|
RIKEN cDNA 1700034O15 gene |
| chr11_-_70255329 | 0.78 |
ENSMUST00000108574.2
ENSMUST00000000329.2 |
Alox12
|
arachidonate 12-lipoxygenase |
| chr1_-_174250976 | 0.78 |
ENSMUST00000061990.4
|
Olfr419
|
olfactory receptor 419 |
| chr2_+_25423234 | 0.78 |
ENSMUST00000134259.1
ENSMUST00000100320.4 |
Fut7
|
fucosyltransferase 7 |
| chr3_-_86920830 | 0.78 |
ENSMUST00000029719.8
|
Dclk2
|
doublecortin-like kinase 2 |
| chr11_+_57645417 | 0.78 |
ENSMUST00000066987.7
ENSMUST00000108846.1 |
Galnt10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 |
| chr19_+_53310495 | 0.77 |
ENSMUST00000003870.7
|
Mxi1
|
Max interacting protein 1 |
| chr13_+_3837757 | 0.77 |
ENSMUST00000042219.4
|
Calm4
|
calmodulin 4 |
| chr7_+_30712209 | 0.77 |
ENSMUST00000005692.6
ENSMUST00000170371.1 |
Atp4a
|
ATPase, H+/K+ exchanging, gastric, alpha polypeptide |
| chr14_+_66344369 | 0.77 |
ENSMUST00000118426.1
ENSMUST00000121955.1 ENSMUST00000120229.1 ENSMUST00000134440.1 |
Stmn4
|
stathmin-like 4 |
| chr2_+_103970221 | 0.76 |
ENSMUST00000111140.2
ENSMUST00000111139.2 |
Lmo2
|
LIM domain only 2 |
| chr11_+_49293297 | 0.76 |
ENSMUST00000054866.3
|
Olfr1392
|
olfactory receptor 1392 |
| chr3_-_95307132 | 0.76 |
ENSMUST00000015846.2
|
Anxa9
|
annexin A9 |
| chr17_+_36898110 | 0.76 |
ENSMUST00000078438.4
|
Trim31
|
tripartite motif-containing 31 |
| chrX_+_164506320 | 0.76 |
ENSMUST00000033756.2
|
Asb9
|
ankyrin repeat and SOCS box-containing 9 |
| chr7_+_100494044 | 0.75 |
ENSMUST00000153287.1
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr13_-_44455303 | 0.74 |
ENSMUST00000183264.1
|
Gm27007
|
predicted gene, 27007 |
| chr5_+_31054766 | 0.74 |
ENSMUST00000013773.5
ENSMUST00000114646.1 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr2_-_25239850 | 0.74 |
ENSMUST00000043379.3
|
2310002J15Rik
|
RIKEN cDNA 2310002J15 gene |
| chr7_-_105574324 | 0.74 |
ENSMUST00000081165.7
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr9_+_107950952 | 0.74 |
ENSMUST00000049348.3
|
Traip
|
TRAF-interacting protein |
| chr13_+_23574381 | 0.74 |
ENSMUST00000090776.4
|
Hist1h2ad
|
histone cluster 1, H2ad |
| chr6_-_56901870 | 0.74 |
ENSMUST00000101367.2
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr15_+_103009479 | 0.73 |
ENSMUST00000001711.4
|
Hoxc6
|
homeobox C6 |
| chr14_+_58893465 | 0.73 |
ENSMUST00000079960.1
|
Rpl13-ps3
|
ribosomal protein L13, pseudogene 3 |
| chr15_-_98881255 | 0.73 |
ENSMUST00000024518.9
|
Rhebl1
|
Ras homolog enriched in brain like 1 |
| chr4_-_118809814 | 0.73 |
ENSMUST00000105035.1
ENSMUST00000084313.3 |
Olfr1335
|
olfactory receptor 1335 |
| chr17_-_33718591 | 0.73 |
ENSMUST00000174040.1
ENSMUST00000173015.1 ENSMUST00000066121.6 ENSMUST00000172767.1 ENSMUST00000173329.1 |
March2
|
membrane-associated ring finger (C3HC4) 2 |
| chr10_-_80813486 | 0.72 |
ENSMUST00000181039.1
ENSMUST00000180438.1 |
Jsrp1
|
junctional sarcoplasmic reticulum protein 1 |
| chr8_-_21096050 | 0.72 |
ENSMUST00000077452.3
|
Gm14851
|
predicted gene 14851 |
| chr17_-_43543639 | 0.72 |
ENSMUST00000178772.1
|
Ankrd66
|
ankyrin repeat domain 66 |
| chr1_+_146420434 | 0.72 |
ENSMUST00000163646.1
|
Gm5263
|
predicted gene 5263 |
| chr4_-_92191749 | 0.72 |
ENSMUST00000123179.1
|
Gm12666
|
predicted gene 12666 |
| chr5_+_136967859 | 0.71 |
ENSMUST00000001790.5
|
Cldn15
|
claudin 15 |
| chr5_+_90772435 | 0.71 |
ENSMUST00000031320.6
|
Pf4
|
platelet factor 4 |
| chr11_-_100207507 | 0.71 |
ENSMUST00000007272.7
|
Krt14
|
keratin 14 |
| chr15_-_101680281 | 0.71 |
ENSMUST00000023786.5
|
Krt6b
|
keratin 6B |
| chr13_+_18948344 | 0.70 |
ENSMUST00000003345.7
|
Amph
|
amphiphysin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gata3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 16.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.9 | 3.4 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.7 | 2.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.6 | 1.9 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.5 | 1.5 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.5 | 1.4 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.4 | 1.3 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.4 | 1.3 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.4 | 0.4 | GO:0060197 | cloacal septation(GO:0060197) |
| 0.4 | 2.5 | GO:0032796 | uropod organization(GO:0032796) |
| 0.4 | 2.4 | GO:0051790 | acetate metabolic process(GO:0006083) short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.4 | 1.9 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.4 | 1.2 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.4 | 4.5 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.4 | 1.1 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.4 | 3.9 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.3 | 1.8 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.3 | 1.2 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.3 | 0.9 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.3 | 0.9 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.3 | 0.8 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.3 | 3.8 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.3 | 0.5 | GO:0060031 | mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.3 | 1.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.3 | 2.1 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) |
| 0.3 | 1.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.3 | 1.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.2 | 1.0 | GO:0036233 | glycine import(GO:0036233) |
| 0.2 | 1.2 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) |
| 0.2 | 1.4 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.2 | 0.9 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 0.9 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.2 | 1.1 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.2 | 0.6 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.2 | 1.3 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.2 | 1.5 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.2 | 0.6 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.2 | 0.8 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.2 | 1.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 0.6 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 0.6 | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 0.6 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 0.2 | 0.6 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.2 | 1.0 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.2 | 1.0 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.2 | 0.6 | GO:1901738 | regulation of vitamin A metabolic process(GO:1901738) |
| 0.2 | 0.8 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.2 | 1.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.2 | 0.2 | GO:0070433 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.2 | 0.7 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.2 | 0.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.2 | 1.4 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.2 | 0.5 | GO:0036088 | D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.2 | 0.5 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.2 | 2.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 | 0.8 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 | 0.8 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.2 | 0.8 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.2 | 1.1 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 1.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 0.4 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 0.6 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.1 | 2.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.7 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 0.4 | GO:2000371 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.1 | 0.4 | GO:0031508 | chromatin remodeling at centromere(GO:0031055) pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 3.7 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 0.1 | GO:0072244 | metanephric glomerular epithelium development(GO:0072244) |
| 0.1 | 0.7 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.1 | 1.3 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.4 | GO:0032910 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.1 | 1.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.5 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 0.8 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.1 | 0.3 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.1 | 1.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.4 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.1 | 0.6 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.1 | 2.8 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 1.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.8 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 1.6 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.4 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.1 | 0.6 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.1 | 0.4 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 0.1 | 0.5 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 2.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.1 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.1 | 0.8 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 0.3 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 1.7 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.1 | 0.6 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.1 | 1.4 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.1 | 0.3 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.1 | 0.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.4 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.5 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.4 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 1.0 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.1 | 0.6 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.1 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.1 | 0.8 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 1.3 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.1 | 0.5 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.1 | 0.1 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.4 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.1 | 0.3 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.1 | 2.1 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.4 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.1 | 0.4 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.1 | 0.8 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 2.9 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.4 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.4 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.1 | 0.4 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.3 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.4 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.3 | GO:0060032 | notochord regression(GO:0060032) |
| 0.1 | 1.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.6 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 | 1.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.3 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.3 | GO:0045588 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.1 | 0.5 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 0.4 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.1 | 0.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.6 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.6 | GO:0043585 | nose morphogenesis(GO:0043585) alveolar primary septum development(GO:0061143) |
| 0.1 | 0.8 | GO:1902571 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 0.1 | GO:0060459 | left lung development(GO:0060459) left lung morphogenesis(GO:0060460) |
| 0.1 | 0.2 | GO:0048627 | myoblast development(GO:0048627) |
| 0.1 | 6.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.1 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.1 | 0.7 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.8 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.1 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.2 | GO:2000978 | auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.1 | 0.7 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.1 | 15.7 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 0.2 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.1 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 0.4 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 1.3 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 0.2 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.1 | 0.4 | GO:2000323 | circadian regulation of translation(GO:0097167) negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 0.4 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.2 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 0.2 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 0.1 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.2 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 | 2.3 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.2 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 | 0.3 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.1 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.2 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.1 | 0.3 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.1 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.6 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.3 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 | 0.6 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.4 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 0.5 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.3 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.7 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.4 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.4 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.2 | GO:0006113 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.1 | 0.5 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.1 | 0.1 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.1 | 0.6 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.2 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 | 0.5 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 0.1 | GO:0044557 | relaxation of smooth muscle(GO:0044557) |
| 0.1 | 0.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.7 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.8 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.1 | 0.8 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.1 | 0.2 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.4 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 1.0 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.2 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 0.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.4 | GO:1901724 | positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 0.1 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 1.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.2 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.1 | 0.3 | GO:0021592 | fourth ventricle development(GO:0021592) initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.9 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.4 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.6 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 1.3 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.1 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.0 | 0.9 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.3 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.7 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 1.3 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.4 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 0.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.7 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.5 | GO:0043084 | penile erection(GO:0043084) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.0 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.0 | 0.5 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.2 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.2 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.2 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.6 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.1 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.0 | 1.0 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 4.2 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.8 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.4 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.0 | 0.3 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.0 | 0.3 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 1.2 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.8 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.1 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.1 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 | 1.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.3 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.1 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.0 | 0.3 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:0045814 | negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.0 | 0.2 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.2 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.4 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.5 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.1 | GO:0033575 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 | 0.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 1.4 | GO:0009712 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 1.5 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.3 | GO:1904871 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 1.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.3 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.9 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) |
| 0.0 | 0.5 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 | 1.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.9 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.4 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 0.2 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.0 | 0.1 | GO:1903093 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.1 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.1 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.0 | 0.2 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.1 | GO:0060618 | nipple development(GO:0060618) |
| 0.0 | 0.2 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.0 | 0.4 | GO:0044036 | cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 0.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.3 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 1.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.1 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.1 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.0 | 0.2 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.1 | GO:0043308 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) cytosine metabolic process(GO:0019858) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.2 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 1.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.2 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.2 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.5 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.5 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.1 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.0 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.6 | GO:0051567 | histone H3-K9 methylation(GO:0051567) |
| 0.0 | 0.2 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.0 | 0.2 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.2 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.0 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.4 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.8 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0061038 | uterus morphogenesis(GO:0061038) |
| 0.0 | 0.4 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.4 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.0 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.4 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.9 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.2 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.2 | GO:0036309 | T-tubule organization(GO:0033292) protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 | 0.1 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.4 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 1.4 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.0 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.7 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.0 | 0.1 | GO:0032700 | antifungal humoral response(GO:0019732) negative regulation of interleukin-17 production(GO:0032700) |
| 0.0 | 0.1 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.3 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.3 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.3 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.2 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.2 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.0 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 1.0 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0033127 | regulation of histone phosphorylation(GO:0033127) |
| 0.0 | 0.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.2 | GO:0032288 | myelin assembly(GO:0032288) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 18.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.5 | 2.1 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.5 | 1.9 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.4 | 1.3 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.4 | 1.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.4 | 1.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.3 | 2.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.3 | 1.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.2 | 1.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 1.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 1.3 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.2 | 0.6 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.2 | 0.6 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.2 | 6.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 1.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 0.7 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 0.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.7 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 0.9 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 11.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.4 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.1 | 2.4 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.9 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 2.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.3 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 0.5 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.4 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 3.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.3 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 7.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 2.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.5 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.3 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 0.3 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.1 | 0.2 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.5 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 3.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.3 | GO:0090537 | CERF complex(GO:0090537) |
| 0.1 | 0.4 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 0.1 | 0.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 2.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.2 | GO:0071564 | SWI/SNF complex(GO:0016514) npBAF complex(GO:0071564) |
| 0.1 | 0.2 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.2 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.6 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 0.7 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.7 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.2 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.6 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.8 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 1.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 1.2 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 3.1 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 1.0 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 2.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 4.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 1.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 1.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.7 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 1.0 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.4 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.0 | 0.6 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 10.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 1.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 1.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 2.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0032040 | 90S preribosome(GO:0030686) small-subunit processome(GO:0032040) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 3.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 16.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.8 | 2.4 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.8 | 4.8 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.6 | 1.3 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.4 | 1.7 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.4 | 1.3 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.4 | 1.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.4 | 3.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.4 | 1.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 1.0 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.3 | 2.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.3 | 0.9 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.3 | 1.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.3 | 1.7 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.3 | 0.8 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.2 | 1.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.2 | 0.7 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.2 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 0.9 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.2 | 0.8 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.2 | 1.0 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.2 | 0.6 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.2 | 0.8 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.2 | 0.7 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.2 | 0.5 | GO:1902271 | lithocholic acid binding(GO:1902121) D3 vitamins binding(GO:1902271) |
| 0.2 | 1.6 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.2 | 2.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 1.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 0.5 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 0.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 0.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.4 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.1 | 1.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.4 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 0.7 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 2.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.8 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 1.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.5 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 0.4 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.6 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.9 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 19.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.7 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.7 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.6 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 0.6 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 0.3 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.7 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 2.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.3 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.1 | 0.3 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.1 | 0.9 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 1.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 1.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.8 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.3 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 1.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 1.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.6 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 3.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.6 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.4 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 0.7 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 3.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.5 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.2 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.1 | 0.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.4 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.5 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.9 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.3 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.2 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.1 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.0 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.2 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.1 | 0.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.2 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.1 | 0.4 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 1.3 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.4 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 5.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.2 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 2.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 2.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.3 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 7.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 1.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.2 | GO:0008443 | phosphofructokinase activity(GO:0008443) |
| 0.0 | 0.6 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.9 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.5 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0031687 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) A2A adenosine receptor binding(GO:0031687) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 2.6 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.4 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 1.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 1.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.3 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.1 | GO:0038100 | nodal binding(GO:0038100) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 3.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.4 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 4.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.1 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 8.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.3 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.8 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0008527 | taste receptor activity(GO:0008527) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 3.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.9 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 2.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.9 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 7.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 3.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 3.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 0.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 3.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 2.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 9.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 3.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 4.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 2.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 4.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 1.1 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.6 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.7 | REACTOME REGULATION OF MITOTIC CELL CYCLE | Genes involved in Regulation of mitotic cell cycle |
| 0.0 | 1.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.0 | 0.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 1.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |