Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
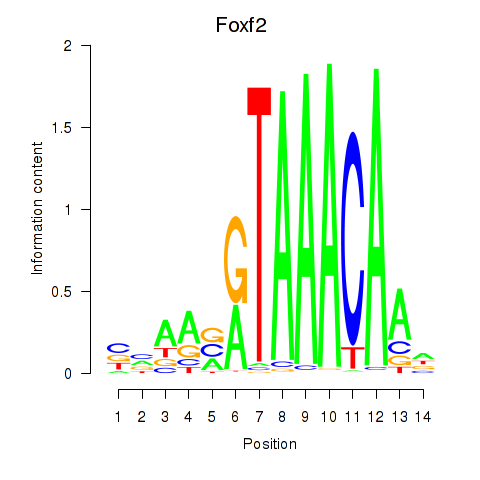
Results for Foxf2
Z-value: 1.66
Transcription factors associated with Foxf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxf2
|
ENSMUSG00000038402.2 | forkhead box F2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxf2 | mm10_v2_chr13_+_31625802_31625816 | 0.39 | 1.8e-02 | Click! |
Activity profile of Foxf2 motif
Sorted Z-values of Foxf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_106167564 | 11.06 |
ENSMUST00000063062.8
|
Chi3l3
|
chitinase 3-like 3 |
| chr8_+_70373541 | 8.39 |
ENSMUST00000003659.7
|
Comp
|
cartilage oligomeric matrix protein |
| chr8_-_85380964 | 8.38 |
ENSMUST00000122452.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr4_-_87806296 | 7.48 |
ENSMUST00000126353.1
ENSMUST00000149357.1 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr4_-_87806276 | 5.67 |
ENSMUST00000148059.1
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr2_-_84822546 | 5.50 |
ENSMUST00000028471.5
|
Smtnl1
|
smoothelin-like 1 |
| chr10_+_88091070 | 5.22 |
ENSMUST00000048621.7
|
Pmch
|
pro-melanin-concentrating hormone |
| chr13_-_23465872 | 5.22 |
ENSMUST00000041674.7
|
Btn1a1
|
butyrophilin, subfamily 1, member A1 |
| chr10_+_115817247 | 5.18 |
ENSMUST00000035563.7
ENSMUST00000080630.3 ENSMUST00000179196.1 |
Tspan8
|
tetraspanin 8 |
| chr15_+_79348061 | 4.41 |
ENSMUST00000163691.1
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr2_-_118728345 | 4.40 |
ENSMUST00000159756.1
|
Plcb2
|
phospholipase C, beta 2 |
| chr2_-_126500631 | 4.31 |
ENSMUST00000129187.1
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr17_+_47505117 | 4.12 |
ENSMUST00000183044.1
ENSMUST00000037333.10 |
Ccnd3
|
cyclin D3 |
| chr15_+_79347534 | 3.99 |
ENSMUST00000096350.3
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr17_+_47505149 | 3.96 |
ENSMUST00000183177.1
ENSMUST00000182848.1 |
Ccnd3
|
cyclin D3 |
| chr2_-_118728430 | 3.83 |
ENSMUST00000102524.1
|
Plcb2
|
phospholipase C, beta 2 |
| chr2_-_170406501 | 3.77 |
ENSMUST00000154650.1
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr17_+_47505211 | 3.71 |
ENSMUST00000182935.1
ENSMUST00000182506.1 |
Ccnd3
|
cyclin D3 |
| chr9_-_58741543 | 3.65 |
ENSMUST00000098674.4
|
2410076I21Rik
|
RIKEN cDNA 2410076I21 gene |
| chr13_-_23465895 | 3.63 |
ENSMUST00000110434.1
|
Btn1a1
|
butyrophilin, subfamily 1, member A1 |
| chr15_-_66969616 | 3.41 |
ENSMUST00000170903.1
ENSMUST00000166420.1 ENSMUST00000005256.6 ENSMUST00000164070.1 |
Ndrg1
|
N-myc downstream regulated gene 1 |
| chr8_+_86745679 | 2.92 |
ENSMUST00000098532.2
|
Gm10638
|
predicted gene 10638 |
| chr10_+_37139558 | 2.83 |
ENSMUST00000062667.3
|
5930403N24Rik
|
RIKEN cDNA 5930403N24 gene |
| chr12_-_111980751 | 2.79 |
ENSMUST00000170525.1
|
BC048943
|
cDNA sequence BC048943 |
| chr14_+_55854115 | 2.71 |
ENSMUST00000168479.1
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr7_-_98145472 | 2.49 |
ENSMUST00000098281.2
|
Omp
|
olfactory marker protein |
| chr2_-_163645125 | 2.44 |
ENSMUST00000017851.3
|
Serinc3
|
serine incorporator 3 |
| chr5_+_118560719 | 2.42 |
ENSMUST00000100816.4
|
Med13l
|
mediator complex subunit 13-like |
| chr2_-_126499839 | 2.42 |
ENSMUST00000040128.5
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr15_-_58324161 | 2.39 |
ENSMUST00000022985.1
|
Klhl38
|
kelch-like 38 |
| chr8_+_93810832 | 2.35 |
ENSMUST00000034198.8
ENSMUST00000125716.1 |
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chrX_+_163911401 | 2.32 |
ENSMUST00000140845.1
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr7_+_30291659 | 2.30 |
ENSMUST00000014065.8
ENSMUST00000150892.1 ENSMUST00000126216.1 |
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr14_+_55853997 | 2.30 |
ENSMUST00000100529.3
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr4_+_101507947 | 2.15 |
ENSMUST00000149047.1
ENSMUST00000106929.3 |
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr19_+_53329413 | 1.98 |
ENSMUST00000025998.7
|
Mxi1
|
Max interacting protein 1 |
| chr18_-_88927447 | 1.93 |
ENSMUST00000147313.1
|
Socs6
|
suppressor of cytokine signaling 6 |
| chr5_-_115098295 | 1.87 |
ENSMUST00000100848.2
|
Gm10401
|
predicted gene 10401 |
| chr7_+_30291941 | 1.85 |
ENSMUST00000144508.1
|
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr16_-_29946499 | 1.85 |
ENSMUST00000181968.1
|
Gm26569
|
predicted gene, 26569 |
| chr11_+_69095217 | 1.84 |
ENSMUST00000101004.2
|
Per1
|
period circadian clock 1 |
| chr15_-_50882806 | 1.75 |
ENSMUST00000184885.1
|
Trps1
|
trichorhinophalangeal syndrome I (human) |
| chr3_-_27896360 | 1.74 |
ENSMUST00000058077.3
|
Tmem212
|
transmembrane protein 212 |
| chr10_-_37138863 | 1.74 |
ENSMUST00000092584.5
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr5_+_86071734 | 1.71 |
ENSMUST00000031171.7
|
Stap1
|
signal transducing adaptor family member 1 |
| chr3_-_93015669 | 1.68 |
ENSMUST00000107301.1
ENSMUST00000029521.4 |
Crct1
|
cysteine-rich C-terminal 1 |
| chr9_+_70678950 | 1.66 |
ENSMUST00000067880.6
|
Adam10
|
a disintegrin and metallopeptidase domain 10 |
| chr1_+_43445736 | 1.56 |
ENSMUST00000086421.5
ENSMUST00000114744.1 |
Nck2
|
non-catalytic region of tyrosine kinase adaptor protein 2 |
| chr13_+_44840686 | 1.54 |
ENSMUST00000173906.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr7_-_142372210 | 1.54 |
ENSMUST00000084412.5
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr13_-_95478655 | 1.49 |
ENSMUST00000022186.3
|
S100z
|
S100 calcium binding protein, zeta |
| chr19_+_23723279 | 1.45 |
ENSMUST00000067077.1
|
Gm9938
|
predicted gene 9938 |
| chr2_-_168601620 | 1.43 |
ENSMUST00000171689.1
ENSMUST00000137451.1 |
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
| chr12_+_69296676 | 1.39 |
ENSMUST00000021362.4
|
Klhdc2
|
kelch domain containing 2 |
| chr19_+_45149833 | 1.34 |
ENSMUST00000026236.9
|
Tlx1
|
T cell leukemia, homeobox 1 |
| chr4_+_101507855 | 1.26 |
ENSMUST00000038207.5
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr2_+_26973416 | 1.24 |
ENSMUST00000014996.7
ENSMUST00000102891.3 |
Adamts13
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 13 |
| chr12_+_3807076 | 1.23 |
ENSMUST00000174817.1
|
Dnmt3a
|
DNA methyltransferase 3A |
| chr17_+_29090969 | 1.20 |
ENSMUST00000119901.1
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr13_+_55445301 | 1.17 |
ENSMUST00000001115.8
ENSMUST00000099482.3 |
Grk6
|
G protein-coupled receptor kinase 6 |
| chrX_-_10216918 | 1.15 |
ENSMUST00000072393.2
ENSMUST00000044598.6 ENSMUST00000073392.4 ENSMUST00000115533.1 ENSMUST00000115532.1 |
Rpgr
|
retinitis pigmentosa GTPase regulator |
| chr3_+_53488677 | 1.15 |
ENSMUST00000029307.3
|
Stoml3
|
stomatin (Epb7.2)-like 3 |
| chr4_+_5724304 | 1.13 |
ENSMUST00000108380.1
|
Fam110b
|
family with sequence similarity 110, member B |
| chr1_+_179546303 | 1.11 |
ENSMUST00000040706.8
|
Cnst
|
consortin, connexin sorting protein |
| chr12_+_3807017 | 1.09 |
ENSMUST00000020991.8
ENSMUST00000172509.1 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr4_+_155791172 | 1.05 |
ENSMUST00000105593.1
|
Ankrd65
|
ankyrin repeat domain 65 |
| chr2_+_112265809 | 1.00 |
ENSMUST00000110991.2
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr18_-_47333311 | 0.96 |
ENSMUST00000126684.1
ENSMUST00000156422.1 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr12_-_15816762 | 0.95 |
ENSMUST00000020922.7
|
Trib2
|
tribbles homolog 2 (Drosophila) |
| chr7_-_30559828 | 0.92 |
ENSMUST00000108164.1
|
Lin37
|
lin-37 homolog (C. elegans) |
| chr14_+_26514554 | 0.92 |
ENSMUST00000104927.1
|
Gm2178
|
predicted gene 2178 |
| chr6_+_147032528 | 0.91 |
ENSMUST00000036194.4
|
Rep15
|
RAB15 effector protein |
| chr4_-_55532453 | 0.90 |
ENSMUST00000132746.1
ENSMUST00000107619.2 |
Klf4
|
Kruppel-like factor 4 (gut) |
| chr5_+_62813823 | 0.90 |
ENSMUST00000170704.1
|
Gm17384
|
predicted gene, 17384 |
| chr18_-_65393844 | 0.89 |
ENSMUST00000035548.8
|
Alpk2
|
alpha-kinase 2 |
| chr11_-_54956047 | 0.87 |
ENSMUST00000155316.1
ENSMUST00000108889.3 ENSMUST00000126703.1 |
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr5_-_6876523 | 0.85 |
ENSMUST00000164784.1
|
Zfp804b
|
zinc finger protein 804B |
| chr4_-_82885148 | 0.81 |
ENSMUST00000048430.3
|
Cer1
|
cerberus 1 homolog (Xenopus laevis) |
| chr15_-_79441802 | 0.80 |
ENSMUST00000122044.1
ENSMUST00000135519.1 |
Csnk1e
|
casein kinase 1, epsilon |
| chr16_-_4880284 | 0.77 |
ENSMUST00000037843.6
|
Ubald1
|
UBA-like domain containing 1 |
| chr10_+_25408346 | 0.77 |
ENSMUST00000092645.6
|
Epb4.1l2
|
erythrocyte protein band 4.1-like 2 |
| chr2_-_73312701 | 0.76 |
ENSMUST00000058615.9
|
Cir1
|
corepressor interacting with RBPJ, 1 |
| chr15_+_54410755 | 0.76 |
ENSMUST00000036737.3
|
Colec10
|
collectin sub-family member 10 |
| chr13_+_22087878 | 0.76 |
ENSMUST00000091739.1
|
Vmn1r188
|
vomeronasal 1 receptor 188 |
| chr1_-_39720989 | 0.74 |
ENSMUST00000151913.1
|
Rfx8
|
regulatory factor X 8 |
| chr6_-_57844493 | 0.72 |
ENSMUST00000081186.3
|
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr8_-_125492710 | 0.70 |
ENSMUST00000108775.1
|
Sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr3_-_37232565 | 0.70 |
ENSMUST00000161015.1
ENSMUST00000029273.1 |
Il21
|
interleukin 21 |
| chr2_-_181578906 | 0.67 |
ENSMUST00000136875.1
|
Uckl1
|
uridine-cytidine kinase 1-like 1 |
| chr10_+_40349265 | 0.67 |
ENSMUST00000044672.4
ENSMUST00000095743.2 |
Cdk19
|
cyclin-dependent kinase 19 |
| chr15_-_66560997 | 0.66 |
ENSMUST00000048372.5
|
Tmem71
|
transmembrane protein 71 |
| chr15_-_79441999 | 0.66 |
ENSMUST00000117786.1
ENSMUST00000120859.1 |
Csnk1e
|
casein kinase 1, epsilon |
| chr14_+_58072686 | 0.65 |
ENSMUST00000022545.7
|
Fgf9
|
fibroblast growth factor 9 |
| chr7_-_44849075 | 0.63 |
ENSMUST00000047085.8
|
Tbc1d17
|
TBC1 domain family, member 17 |
| chr11_-_120990871 | 0.62 |
ENSMUST00000154483.1
|
Csnk1d
|
casein kinase 1, delta |
| chr5_+_107437908 | 0.62 |
ENSMUST00000094541.2
|
Btbd8
|
BTB (POZ) domain containing 8 |
| chr19_-_45783512 | 0.59 |
ENSMUST00000026243.3
|
Mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr10_+_126978690 | 0.58 |
ENSMUST00000105256.2
|
Ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr12_-_112511136 | 0.57 |
ENSMUST00000066791.5
|
Tmem179
|
transmembrane protein 179 |
| chr2_-_60125651 | 0.57 |
ENSMUST00000112550.1
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr9_-_108649349 | 0.56 |
ENSMUST00000013338.8
|
Arih2
|
ariadne homolog 2 (Drosophila) |
| chr7_+_90426312 | 0.53 |
ENSMUST00000061391.7
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr15_+_91838322 | 0.53 |
ENSMUST00000160242.1
ENSMUST00000109276.1 ENSMUST00000088555.3 ENSMUST00000100293.2 ENSMUST00000126508.1 |
Muc19
Smgc
|
mucin 19 submandibular gland protein C |
| chr18_+_55057557 | 0.51 |
ENSMUST00000181765.1
|
Gm4221
|
predicted gene 4221 |
| chr1_+_51289106 | 0.49 |
ENSMUST00000051572.6
|
Sdpr
|
serum deprivation response |
| chr1_+_40681659 | 0.45 |
ENSMUST00000027231.7
|
Slc9a2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2 |
| chr2_-_71367749 | 0.43 |
ENSMUST00000151937.1
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr12_+_71016658 | 0.40 |
ENSMUST00000125125.1
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr7_+_44849216 | 0.37 |
ENSMUST00000054343.8
|
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr10_-_49783259 | 0.35 |
ENSMUST00000105484.3
ENSMUST00000105486.1 ENSMUST00000079751.2 ENSMUST00000105485.1 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr7_+_82175156 | 0.31 |
ENSMUST00000180243.1
|
Sh3gl3
|
SH3-domain GRB2-like 3 |
| chr18_+_34759551 | 0.30 |
ENSMUST00000097622.3
|
Fam53c
|
family with sequence similarity 53, member C |
| chr3_+_65528457 | 0.29 |
ENSMUST00000130705.1
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr16_-_79091078 | 0.27 |
ENSMUST00000023566.4
ENSMUST00000060402.5 |
Tmprss15
|
transmembrane protease, serine 15 |
| chr1_+_34005872 | 0.27 |
ENSMUST00000182296.1
|
Dst
|
dystonin |
| chr6_+_34863130 | 0.25 |
ENSMUST00000074949.3
|
Tmem140
|
transmembrane protein 140 |
| chr3_+_65528404 | 0.23 |
ENSMUST00000047906.3
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr11_+_85311232 | 0.19 |
ENSMUST00000020835.9
|
Ppm1d
|
protein phosphatase 1D magnesium-dependent, delta isoform |
| chr16_-_22439719 | 0.18 |
ENSMUST00000079601.6
|
Etv5
|
ets variant gene 5 |
| chr16_-_22439570 | 0.17 |
ENSMUST00000170393.1
|
Etv5
|
ets variant gene 5 |
| chr5_-_103211251 | 0.13 |
ENSMUST00000060871.5
ENSMUST00000112846.1 ENSMUST00000170792.1 ENSMUST00000112847.2 |
Mapk10
|
mitogen-activated protein kinase 10 |
| chrX_-_79434418 | 0.10 |
ENSMUST00000179788.1
|
Chdc2
|
calponin homology domain containing 2 |
| chr5_+_87925579 | 0.09 |
ENSMUST00000001667.6
ENSMUST00000113267.1 |
Csn3
|
casein kappa |
| chr4_-_118409219 | 0.06 |
ENSMUST00000075406.5
|
Szt2
|
seizure threshold 2 |
| chr7_+_44848991 | 0.05 |
ENSMUST00000107885.1
|
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr9_+_44072196 | 0.05 |
ENSMUST00000176671.1
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr10_-_26373956 | 0.04 |
ENSMUST00000105519.3
ENSMUST00000040219.6 |
L3mbtl3
|
l(3)mbt-like 3 (Drosophila) |
| chr8_-_18741542 | 0.03 |
ENSMUST00000033846.6
|
Angpt2
|
angiopoietin 2 |
| chr3_+_51693771 | 0.01 |
ENSMUST00000099104.2
|
Gm10729
|
predicted gene 10729 |
| chr7_-_4844665 | 0.00 |
ENSMUST00000066041.5
ENSMUST00000172377.1 |
Shisa7
|
shisa homolog 7 (Xenopus laevis) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 1.4 | 8.4 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.8 | 13.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.6 | 8.8 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.6 | 5.0 | GO:0015074 | DNA integration(GO:0015074) |
| 0.6 | 4.2 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.6 | 1.7 | GO:1903972 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.6 | 3.4 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.5 | 11.8 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.5 | 4.0 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.4 | 8.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.4 | 1.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.4 | 1.8 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.4 | 9.2 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.3 | 1.7 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.3 | 0.9 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.3 | 3.4 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.3 | 0.9 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.3 | 1.2 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.3 | 2.3 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.2 | 0.7 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.2 | 0.7 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.2 | 2.1 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.2 | 5.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 1.2 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.1 | 8.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.7 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 | 0.6 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) |
| 0.1 | 1.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.9 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 1.8 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 0.4 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.9 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.1 | 0.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 1.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 2.3 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 6.7 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 1.3 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.5 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 2.3 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 1.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 1.0 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.6 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 1.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 1.1 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.4 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 1.9 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.5 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 2.5 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.6 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.6 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 8.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.6 | 1.7 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.4 | 1.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.3 | 11.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 13.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.2 | 7.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 1.7 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 5.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 2.3 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 2.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 2.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 2.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.9 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 3.4 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 1.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 3.7 | GO:0014069 | postsynaptic density(GO:0014069) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 8.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 1.2 | 8.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.7 | 4.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.6 | 1.7 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.4 | 1.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.4 | 8.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.3 | 2.3 | GO:0051430 | G-protein coupled serotonin receptor binding(GO:0031821) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 1.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.3 | 2.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.3 | 5.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 0.8 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.3 | 12.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 0.7 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.2 | 6.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 5.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 0.9 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.2 | 2.4 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.7 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 1.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 3.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.9 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 1.3 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 1.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 1.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 2.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 2.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.9 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 2.3 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 1.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.8 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 11.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 3.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 0.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.6 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 8.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 3.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 10.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 13.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.2 | 2.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 1.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 3.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 6.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.9 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 8.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 3.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 3.9 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.1 | 2.3 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 4.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |