Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
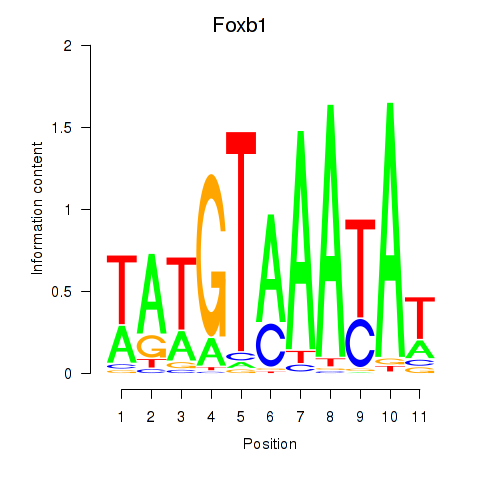
Results for Foxb1
Z-value: 1.25
Transcription factors associated with Foxb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxb1
|
ENSMUSG00000059246.4 | forkhead box B1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxb1 | mm10_v2_chr9_-_69760924_69760940 | -0.66 | 1.2e-05 | Click! |
Activity profile of Foxb1 motif
Sorted Z-values of Foxb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_127898515 | 9.37 |
ENSMUST00000047134.7
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr19_+_20601958 | 6.17 |
ENSMUST00000087638.3
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr19_-_20727533 | 4.53 |
ENSMUST00000025656.3
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr7_+_119900099 | 3.76 |
ENSMUST00000106516.1
|
Lyrm1
|
LYR motif containing 1 |
| chr2_-_94264745 | 2.99 |
ENSMUST00000134563.1
|
E530001K10Rik
|
RIKEN cDNA E530001K10 gene |
| chr2_+_58754910 | 2.87 |
ENSMUST00000059102.6
|
Upp2
|
uridine phosphorylase 2 |
| chr2_+_58755177 | 2.78 |
ENSMUST00000102755.3
|
Upp2
|
uridine phosphorylase 2 |
| chr3_-_95904683 | 2.59 |
ENSMUST00000147962.1
ENSMUST00000036181.8 |
Car14
|
carbonic anhydrase 14 |
| chr12_-_84450944 | 2.42 |
ENSMUST00000085192.5
|
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr3_-_146596588 | 2.37 |
ENSMUST00000029836.4
|
Dnase2b
|
deoxyribonuclease II beta |
| chr13_-_12464925 | 2.21 |
ENSMUST00000124888.1
|
Lgals8
|
lectin, galactose binding, soluble 8 |
| chr7_-_34655500 | 2.09 |
ENSMUST00000032709.1
|
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chr17_+_12378603 | 2.06 |
ENSMUST00000014578.5
|
Plg
|
plasminogen |
| chr7_+_67655414 | 2.04 |
ENSMUST00000107470.1
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr3_+_96245530 | 1.81 |
ENSMUST00000074976.6
|
Hist2h2aa1
|
histone cluster 2, H2aa1 |
| chr3_-_96240317 | 1.78 |
ENSMUST00000078756.5
|
Hist2h2aa2
|
histone cluster 2, H2aa2 |
| chr2_+_55437100 | 1.73 |
ENSMUST00000112633.2
ENSMUST00000112632.1 |
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr14_+_48120841 | 1.66 |
ENSMUST00000073150.4
|
Peli2
|
pellino 2 |
| chr5_+_102481546 | 1.66 |
ENSMUST00000112854.1
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr5_-_34187670 | 1.55 |
ENSMUST00000042701.6
ENSMUST00000119171.1 |
Mxd4
|
Max dimerization protein 4 |
| chr6_+_15185203 | 1.50 |
ENSMUST00000154448.1
|
Foxp2
|
forkhead box P2 |
| chr2_-_94264713 | 1.48 |
ENSMUST00000129661.1
|
E530001K10Rik
|
RIKEN cDNA E530001K10 gene |
| chr17_+_70522083 | 1.43 |
ENSMUST00000148486.1
ENSMUST00000133717.1 |
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr5_+_102481374 | 1.38 |
ENSMUST00000094559.2
ENSMUST00000073302.5 |
Arhgap24
|
Rho GTPase activating protein 24 |
| chr7_+_27591705 | 1.37 |
ENSMUST00000167435.1
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr1_-_179546261 | 1.37 |
ENSMUST00000027769.5
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr7_+_27591513 | 1.36 |
ENSMUST00000108344.2
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chrX_+_150594420 | 1.30 |
ENSMUST00000112713.2
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr2_-_160859601 | 1.28 |
ENSMUST00000103112.1
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chrX_+_101376359 | 1.27 |
ENSMUST00000119080.1
|
Gjb1
|
gap junction protein, beta 1 |
| chr4_+_102430047 | 1.27 |
ENSMUST00000172616.1
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr1_-_170867761 | 1.21 |
ENSMUST00000027974.6
|
Atf6
|
activating transcription factor 6 |
| chr10_+_111506286 | 1.19 |
ENSMUST00000164773.1
|
Phlda1
|
pleckstrin homology-like domain, family A, member 1 |
| chr2_+_137663424 | 1.18 |
ENSMUST00000134833.1
|
Gm14064
|
predicted gene 14064 |
| chr11_+_78465697 | 1.11 |
ENSMUST00000001126.3
|
Slc46a1
|
solute carrier family 46, member 1 |
| chr14_-_55643251 | 1.11 |
ENSMUST00000120041.1
ENSMUST00000121937.1 ENSMUST00000133707.1 ENSMUST00000002391.8 ENSMUST00000121791.1 |
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr9_+_47530173 | 1.11 |
ENSMUST00000114548.1
ENSMUST00000152459.1 ENSMUST00000143026.1 ENSMUST00000085909.2 ENSMUST00000114547.1 ENSMUST00000034581.3 |
Cadm1
|
cell adhesion molecule 1 |
| chr17_+_34969912 | 1.07 |
ENSMUST00000173680.1
|
Gm20481
|
predicted gene 20481 |
| chr7_+_19228334 | 1.06 |
ENSMUST00000063976.8
|
Opa3
|
optic atrophy 3 |
| chr6_+_8948608 | 1.01 |
ENSMUST00000160300.1
|
Nxph1
|
neurexophilin 1 |
| chr10_-_96409038 | 1.00 |
ENSMUST00000179683.1
|
Gm20091
|
predicted gene, 20091 |
| chr1_+_25830657 | 0.98 |
ENSMUST00000064487.1
|
Gm9884
|
predicted gene 9884 |
| chr8_-_45294854 | 0.97 |
ENSMUST00000116473.2
|
Klkb1
|
kallikrein B, plasma 1 |
| chr13_-_18031616 | 0.97 |
ENSMUST00000099736.2
|
Vdac3-ps1
|
voltage-dependent anion channel 3, pseudogene 1 |
| chr4_-_135494499 | 0.96 |
ENSMUST00000105856.2
|
Nipal3
|
NIPA-like domain containing 3 |
| chr15_+_43477213 | 0.93 |
ENSMUST00000022962.6
|
Emc2
|
ER membrane protein complex subunit 2 |
| chr6_+_14901344 | 0.90 |
ENSMUST00000115477.1
|
Foxp2
|
forkhead box P2 |
| chrX_+_57212110 | 0.88 |
ENSMUST00000033466.1
|
Cd40lg
|
CD40 ligand |
| chr5_+_3657172 | 0.84 |
ENSMUST00000115515.1
|
4930511M11Rik
|
RIKEN cDNA 4930511M11 gene |
| chr10_+_60106452 | 0.84 |
ENSMUST00000165024.2
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr13_+_40859768 | 0.81 |
ENSMUST00000110191.2
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr5_+_104170676 | 0.81 |
ENSMUST00000112771.1
|
Dspp
|
dentin sialophosphoprotein |
| chr13_-_83729544 | 0.78 |
ENSMUST00000181705.1
|
Gm26803
|
predicted gene, 26803 |
| chr2_-_134554348 | 0.75 |
ENSMUST00000028704.2
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr4_+_139653538 | 0.73 |
ENSMUST00000030510.7
ENSMUST00000166773.1 |
Tas1r2
|
taste receptor, type 1, member 2 |
| chr19_+_5041337 | 0.72 |
ENSMUST00000116567.2
|
Brms1
|
breast cancer metastasis-suppressor 1 |
| chr1_-_120120138 | 0.72 |
ENSMUST00000112648.1
ENSMUST00000128408.1 |
Dbi
|
diazepam binding inhibitor |
| chrX_+_109095359 | 0.71 |
ENSMUST00000033598.8
|
Sh3bgrl
|
SH3-binding domain glutamic acid-rich protein like |
| chr6_-_123650635 | 0.68 |
ENSMUST00000170808.1
|
Vmn2r22
|
vomeronasal 2, receptor 22 |
| chr6_+_90122643 | 0.68 |
ENSMUST00000174204.1
|
Vmn1r51
|
vomeronasal 1 receptor 51 |
| chr8_+_94037198 | 0.66 |
ENSMUST00000109556.2
ENSMUST00000093301.2 ENSMUST00000060632.7 |
Ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr10_+_38965515 | 0.65 |
ENSMUST00000019992.5
|
Lama4
|
laminin, alpha 4 |
| chr16_-_34095983 | 0.65 |
ENSMUST00000114973.1
ENSMUST00000114964.1 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr8_-_121083085 | 0.65 |
ENSMUST00000182264.1
|
Fendrr
|
Foxf1 adjacent non-coding developmental regulatory RNA |
| chr11_-_98329641 | 0.64 |
ENSMUST00000041685.6
|
Neurod2
|
neurogenic differentiation 2 |
| chr12_-_31950170 | 0.64 |
ENSMUST00000176520.1
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr4_-_135494615 | 0.64 |
ENSMUST00000102549.3
|
Nipal3
|
NIPA-like domain containing 3 |
| chr5_-_51567717 | 0.61 |
ENSMUST00000127135.1
ENSMUST00000151104.1 |
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr5_-_87482258 | 0.57 |
ENSMUST00000079811.6
ENSMUST00000144144.1 |
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr5_-_88527841 | 0.57 |
ENSMUST00000087033.3
|
Igj
|
immunoglobulin joining chain |
| chr10_+_69925954 | 0.57 |
ENSMUST00000181974.1
ENSMUST00000182795.1 ENSMUST00000182437.1 |
Ank3
|
ankyrin 3, epithelial |
| chr5_-_108795352 | 0.56 |
ENSMUST00000004943.1
|
Tmed11
|
transmembrane emp24 protein transport domain containing |
| chr6_-_136922169 | 0.55 |
ENSMUST00000032343.6
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr12_-_31950535 | 0.54 |
ENSMUST00000172314.2
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr10_+_69925766 | 0.53 |
ENSMUST00000182269.1
ENSMUST00000183261.1 ENSMUST00000183074.1 |
Ank3
|
ankyrin 3, epithelial |
| chr12_-_40199315 | 0.53 |
ENSMUST00000095760.2
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr1_-_72536930 | 0.53 |
ENSMUST00000047786.5
|
March4
|
membrane-associated ring finger (C3HC4) 4 |
| chr18_-_43687695 | 0.51 |
ENSMUST00000082254.6
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr10_+_69925800 | 0.50 |
ENSMUST00000182029.1
|
Ank3
|
ankyrin 3, epithelial |
| chr12_-_31950210 | 0.49 |
ENSMUST00000176084.1
ENSMUST00000176103.1 ENSMUST00000167458.2 |
Hbp1
|
high mobility group box transcription factor 1 |
| chr6_+_14901440 | 0.49 |
ENSMUST00000128567.1
|
Foxp2
|
forkhead box P2 |
| chr17_+_70522149 | 0.48 |
ENSMUST00000140728.1
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr14_-_41185188 | 0.47 |
ENSMUST00000077136.3
|
Sftpd
|
surfactant associated protein D |
| chr7_+_24271568 | 0.44 |
ENSMUST00000032696.6
|
Zfp93
|
zinc finger protein 93 |
| chr2_-_147186389 | 0.44 |
ENSMUST00000109970.3
ENSMUST00000067075.5 |
Nkx2-2
|
NK2 homeobox 2 |
| chr1_-_75046639 | 0.44 |
ENSMUST00000152855.1
|
Nhej1
|
nonhomologous end-joining factor 1 |
| chr16_-_44333135 | 0.42 |
ENSMUST00000047446.6
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr4_+_32243733 | 0.41 |
ENSMUST00000165661.1
|
D130062J21Rik
|
RIKEN cDNA D130062J21 gene |
| chr14_-_51256112 | 0.40 |
ENSMUST00000061936.6
|
Ear11
|
eosinophil-associated, ribonuclease A family, member 11 |
| chr5_-_75044562 | 0.39 |
ENSMUST00000075452.5
|
Chic2
|
cysteine-rich hydrophobic domain 2 |
| chr10_-_61979073 | 0.39 |
ENSMUST00000105453.1
ENSMUST00000105452.2 ENSMUST00000105454.2 |
Col13a1
|
collagen, type XIII, alpha 1 |
| chr2_+_30061754 | 0.39 |
ENSMUST00000149578.1
ENSMUST00000102866.3 |
Set
|
SET nuclear oncogene |
| chr6_+_92940572 | 0.38 |
ENSMUST00000181145.1
ENSMUST00000181840.1 |
9530026P05Rik
|
RIKEN cDNA 9530026P05 gene |
| chr16_-_44332925 | 0.38 |
ENSMUST00000136381.1
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr3_-_129804030 | 0.38 |
ENSMUST00000179187.1
|
Lrit3
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3 |
| chr3_-_33082004 | 0.36 |
ENSMUST00000108225.3
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr18_+_36952621 | 0.34 |
ENSMUST00000115661.2
|
Pcdha2
|
protocadherin alpha 2 |
| chr13_+_83738874 | 0.34 |
ENSMUST00000052354.4
|
C130071C03Rik
|
RIKEN cDNA C130071C03 gene |
| chr1_+_81077204 | 0.34 |
ENSMUST00000123720.1
|
Nyap2
|
neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 |
| chr1_-_156034826 | 0.33 |
ENSMUST00000141878.1
ENSMUST00000123705.1 |
Tor1aip1
|
torsin A interacting protein 1 |
| chr9_-_101198999 | 0.31 |
ENSMUST00000066773.7
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr2_-_176917518 | 0.31 |
ENSMUST00000108931.2
|
Gm14296
|
predicted gene 14296 |
| chr1_-_39805311 | 0.31 |
ENSMUST00000171319.2
|
Gm3646
|
predicted gene 3646 |
| chr3_-_146781351 | 0.30 |
ENSMUST00000005164.7
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr4_-_139833524 | 0.29 |
ENSMUST00000030508.7
|
Pax7
|
paired box gene 7 |
| chr4_+_11579647 | 0.29 |
ENSMUST00000180239.1
|
Fsbp
|
fibrinogen silencer binding protein |
| chr10_+_69925484 | 0.29 |
ENSMUST00000182692.1
ENSMUST00000092433.5 |
Ank3
|
ankyrin 3, epithelial |
| chr2_+_181767040 | 0.29 |
ENSMUST00000108756.1
|
Myt1
|
myelin transcription factor 1 |
| chr4_+_109235413 | 0.26 |
ENSMUST00000106628.1
|
Calr4
|
calreticulin 4 |
| chr16_+_17331371 | 0.26 |
ENSMUST00000023450.6
ENSMUST00000161034.1 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chrX_+_119927196 | 0.25 |
ENSMUST00000040961.2
ENSMUST00000113366.1 |
Pabpc5
|
poly(A) binding protein, cytoplasmic 5 |
| chr3_-_155055341 | 0.25 |
ENSMUST00000064076.3
|
Tnni3k
|
TNNI3 interacting kinase |
| chr7_-_109960385 | 0.24 |
ENSMUST00000106722.1
|
Dennd5a
|
DENN/MADD domain containing 5A |
| chr1_+_179960472 | 0.24 |
ENSMUST00000097453.2
ENSMUST00000111117.1 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr2_+_181767283 | 0.20 |
ENSMUST00000108757.2
|
Myt1
|
myelin transcription factor 1 |
| chr2_+_153900374 | 0.18 |
ENSMUST00000088955.5
ENSMUST00000135501.1 |
Bpifb6
|
BPI fold containing family B, member 6 |
| chr4_+_99656299 | 0.18 |
ENSMUST00000087285.3
|
Foxd3
|
forkhead box D3 |
| chr6_+_34863130 | 0.17 |
ENSMUST00000074949.3
|
Tmem140
|
transmembrane protein 140 |
| chr15_-_55548164 | 0.16 |
ENSMUST00000165356.1
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr7_+_29071597 | 0.14 |
ENSMUST00000180926.1
|
Gm26604
|
predicted gene, 26604 |
| chr7_-_109960461 | 0.14 |
ENSMUST00000080437.6
|
Dennd5a
|
DENN/MADD domain containing 5A |
| chr5_-_107687990 | 0.13 |
ENSMUST00000180428.1
|
Gm26692
|
predicted gene, 26692 |
| chr14_+_55222005 | 0.12 |
ENSMUST00000022820.5
|
Dhrs2
|
dehydrogenase/reductase member 2 |
| chr11_+_87699897 | 0.12 |
ENSMUST00000040089.4
|
Rnf43
|
ring finger protein 43 |
| chr19_-_9087945 | 0.11 |
ENSMUST00000025554.2
|
Scgb1a1
|
secretoglobin, family 1A, member 1 (uteroglobin) |
| chr3_-_123690806 | 0.11 |
ENSMUST00000154668.1
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr6_+_34709610 | 0.11 |
ENSMUST00000031775.6
|
Cald1
|
caldesmon 1 |
| chrX_-_43274786 | 0.10 |
ENSMUST00000016294.7
|
Tenm1
|
teneurin transmembrane protein 1 |
| chrX_+_41401128 | 0.10 |
ENSMUST00000115103.2
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
| chr10_+_96616998 | 0.10 |
ENSMUST00000038377.7
|
Btg1
|
B cell translocation gene 1, anti-proliferative |
| chr7_+_103184803 | 0.09 |
ENSMUST00000106893.1
|
Olfr592
|
olfactory receptor 592 |
| chr4_-_103026709 | 0.06 |
ENSMUST00000084382.5
ENSMUST00000106869.2 |
Insl5
|
insulin-like 5 |
| chr4_-_121109199 | 0.06 |
ENSMUST00000106268.3
|
Tmco2
|
transmembrane and coiled-coil domains 2 |
| chr2_-_60722636 | 0.06 |
ENSMUST00000028348.2
ENSMUST00000112517.1 |
Itgb6
|
integrin beta 6 |
| chr5_-_137502402 | 0.02 |
ENSMUST00000111035.1
ENSMUST00000031728.4 |
Pop7
|
processing of precursor 7, ribonuclease P family, (S. cerevisiae) |
| chr1_+_70725715 | 0.01 |
ENSMUST00000053922.5
|
Vwc2l
|
von Willebrand factor C domain-containing protein 2-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.8 | 2.4 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.5 | 2.5 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.5 | 2.7 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.4 | 1.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.4 | 2.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.3 | 3.0 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.3 | 4.5 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.3 | 0.9 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.3 | 2.6 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.3 | 0.8 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 1.7 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 1.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 0.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.2 | 0.8 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.2 | 2.9 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.2 | 1.9 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 0.6 | GO:1990401 | embryonic lung development(GO:1990401) |
| 0.2 | 0.6 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.2 | 1.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 1.7 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.4 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 1.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.7 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 1.9 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 1.3 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 2.4 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.7 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.7 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.9 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.8 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 1.7 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 | 1.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.8 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 1.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 1.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.7 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 0.6 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.4 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.3 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 4.4 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.0 | 0.6 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 3.6 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 1.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.5 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 1.3 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.4 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.4 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.4 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.7 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.5 | 2.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.4 | 1.3 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 2.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.9 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.4 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 1.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.4 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.7 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 1.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.6 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.1 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.7 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 1.1 | 5.7 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.6 | 2.4 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.4 | 9.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.3 | 1.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.3 | 1.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 1.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 0.7 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.2 | 0.8 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 0.8 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.2 | 1.3 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 2.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.2 | 2.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.8 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 1.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 1.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.7 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 1.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.4 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.7 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 2.4 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 4.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 2.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 3.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 2.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 5.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 2.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.7 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 2.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 3.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.1 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.4 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |