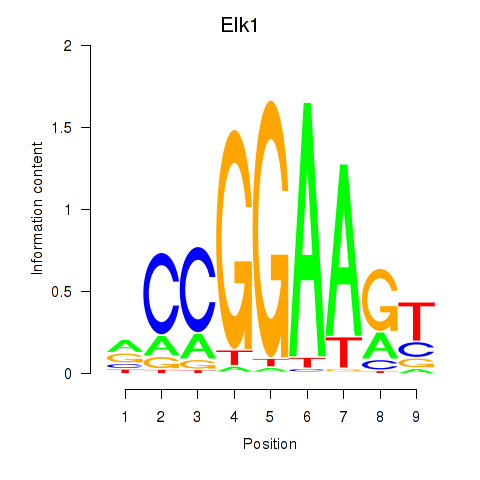
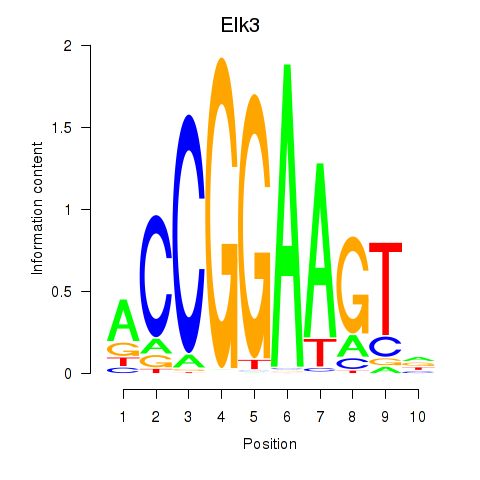
Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
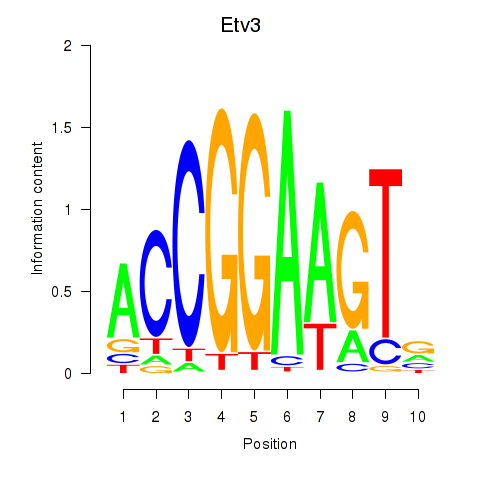
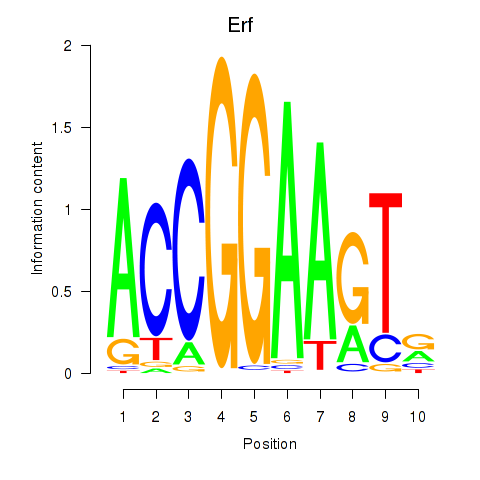
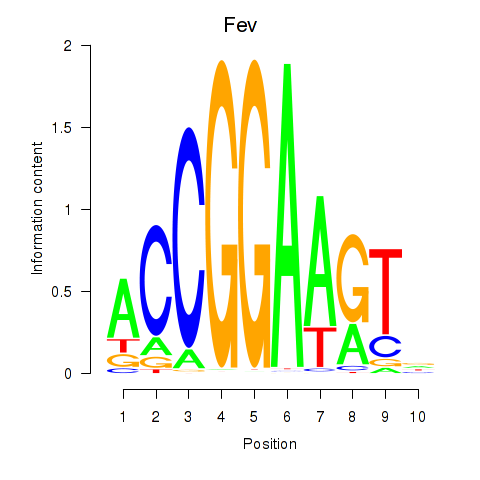
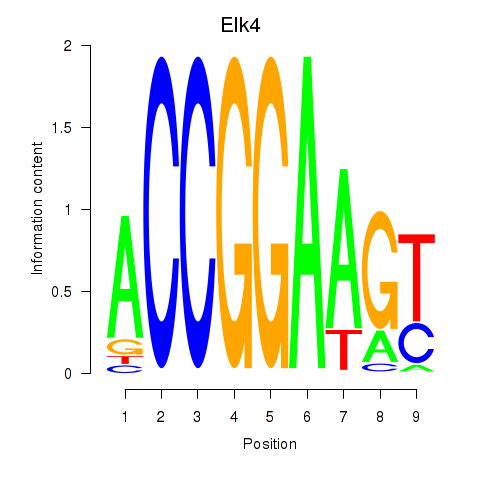
Results for Etv3_Erf_Fev_Elk4_Elk1_Elk3
Z-value: 1.95
Transcription factors associated with Etv3_Erf_Fev_Elk4_Elk1_Elk3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Etv3
|
ENSMUSG00000003382.12 | ets variant 3 |
|
Erf
|
ENSMUSG00000040857.9 | Ets2 repressor factor |
|
Fev
|
ENSMUSG00000055197.4 | FEV transcription factor, ETS family member |
|
Elk4
|
ENSMUSG00000026436.9 | ELK4, member of ETS oncogene family |
|
Elk1
|
ENSMUSG00000009406.7 | ELK1, member of ETS oncogene family |
|
Elk3
|
ENSMUSG00000008398.8 | ELK3, member of ETS oncogene family |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fev | mm10_v2_chr1_-_74885322_74885419 | -0.77 | 3.1e-08 | Click! |
| Elk1 | mm10_v2_chrX_-_20950597_20950613 | -0.59 | 1.5e-04 | Click! |
| Erf | mm10_v2_chr7_-_25250720_25250761 | -0.49 | 2.7e-03 | Click! |
| Elk3 | mm10_v2_chr10_-_93310963_93310989 | -0.44 | 7.4e-03 | Click! |
| Elk4 | mm10_v2_chr1_+_132007606_132007634 | 0.15 | 3.9e-01 | Click! |
| Etv3 | mm10_v2_chr3_+_87525572_87525643 | -0.06 | 7.1e-01 | Click! |
Activity profile of Etv3_Erf_Fev_Elk4_Elk1_Elk3 motif
Sorted Z-values of Etv3_Erf_Fev_Elk4_Elk1_Elk3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_121327672 | 12.37 |
ENSMUST00000159085.1
ENSMUST00000159125.1 ENSMUST00000161818.1 |
Insig2
|
insulin induced gene 2 |
| chr1_-_121328024 | 11.73 |
ENSMUST00000003818.7
|
Insig2
|
insulin induced gene 2 |
| chr1_-_121327734 | 11.71 |
ENSMUST00000160968.1
ENSMUST00000162582.1 |
Insig2
|
insulin induced gene 2 |
| chr1_-_121327776 | 11.35 |
ENSMUST00000160688.1
|
Insig2
|
insulin induced gene 2 |
| chr7_+_87246649 | 7.44 |
ENSMUST00000068829.5
ENSMUST00000032781.7 |
Nox4
|
NADPH oxidase 4 |
| chr11_-_48816936 | 7.26 |
ENSMUST00000140800.1
|
Trim41
|
tripartite motif-containing 41 |
| chr10_-_40025253 | 7.16 |
ENSMUST00000163705.2
|
AI317395
|
expressed sequence AI317395 |
| chr5_-_151369172 | 6.75 |
ENSMUST00000067770.3
|
D730045B01Rik
|
RIKEN cDNA D730045B01 gene |
| chr7_-_79743034 | 6.56 |
ENSMUST00000032761.7
|
Pex11a
|
peroxisomal biogenesis factor 11 alpha |
| chr9_-_65908676 | 6.23 |
ENSMUST00000119245.1
ENSMUST00000134338.1 ENSMUST00000179395.1 |
Trip4
|
thyroid hormone receptor interactor 4 |
| chr11_-_48817332 | 6.14 |
ENSMUST00000047145.7
|
Trim41
|
tripartite motif-containing 41 |
| chr5_+_151368683 | 5.63 |
ENSMUST00000181114.1
ENSMUST00000181555.1 |
1700028E10Rik
|
RIKEN cDNA 1700028E10 gene |
| chr2_-_12419456 | 5.52 |
ENSMUST00000154899.1
ENSMUST00000028105.6 |
Fam188a
|
family with sequence similarity 188, member A |
| chr11_+_87592145 | 5.09 |
ENSMUST00000103179.3
ENSMUST00000092802.5 ENSMUST00000146871.1 |
Mtmr4
|
myotubularin related protein 4 |
| chr16_-_87432597 | 4.94 |
ENSMUST00000039449.7
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr10_-_95324072 | 4.89 |
ENSMUST00000053594.5
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr9_-_22002599 | 4.83 |
ENSMUST00000115336.2
ENSMUST00000044926.5 |
Ccdc151
|
coiled-coil domain containing 151 |
| chr13_-_90089513 | 4.67 |
ENSMUST00000160232.1
|
Xrcc4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr2_-_73386396 | 4.56 |
ENSMUST00000112044.1
ENSMUST00000112043.1 ENSMUST00000076463.5 |
Gpr155
|
G protein-coupled receptor 155 |
| chr2_-_12419387 | 4.53 |
ENSMUST00000124515.1
|
Fam188a
|
family with sequence similarity 188, member A |
| chr17_-_59013264 | 4.31 |
ENSMUST00000174122.1
ENSMUST00000025065.5 |
Nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr8_-_25785154 | 4.28 |
ENSMUST00000038498.8
|
Bag4
|
BCL2-associated athanogene 4 |
| chr13_-_90089556 | 4.04 |
ENSMUST00000022115.7
|
Xrcc4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr7_+_79743142 | 3.98 |
ENSMUST00000035622.7
|
Wdr93
|
WD repeat domain 93 |
| chr11_-_20112876 | 3.95 |
ENSMUST00000000137.7
|
Actr2
|
ARP2 actin-related protein 2 |
| chr9_+_110476985 | 3.91 |
ENSMUST00000084948.4
ENSMUST00000061155.6 ENSMUST00000140686.1 ENSMUST00000084952.5 |
Kif9
|
kinesin family member 9 |
| chr9_+_45055211 | 3.85 |
ENSMUST00000114663.2
|
Mpzl3
|
myelin protein zero-like 3 |
| chr5_-_9161692 | 3.84 |
ENSMUST00000183973.1
ENSMUST00000184372.1 ENSMUST00000095017.4 ENSMUST00000071921.6 |
Dmtf1
|
cyclin D binding myb-like transcription factor 1 |
| chr9_-_44965519 | 3.79 |
ENSMUST00000125642.1
ENSMUST00000117506.1 ENSMUST00000117549.1 |
Ube4a
|
ubiquitination factor E4A, UFD2 homolog (S. cerevisiae) |
| chr19_-_29047847 | 3.71 |
ENSMUST00000025696.4
|
Ak3
|
adenylate kinase 3 |
| chr16_+_3872368 | 3.68 |
ENSMUST00000151988.1
|
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr7_+_51878967 | 3.66 |
ENSMUST00000051912.6
|
Gas2
|
growth arrest specific 2 |
| chr1_-_179546261 | 3.63 |
ENSMUST00000027769.5
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr9_-_106476590 | 3.56 |
ENSMUST00000112479.2
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr6_+_8259288 | 3.51 |
ENSMUST00000159335.1
|
Gm16039
|
predicted gene 16039 |
| chr2_+_73312601 | 3.49 |
ENSMUST00000090811.4
ENSMUST00000112050.1 |
Scrn3
|
secernin 3 |
| chr9_-_105495130 | 3.44 |
ENSMUST00000038118.7
|
Atp2c1
|
ATPase, Ca++-sequestering |
| chrX_-_169320273 | 3.41 |
ENSMUST00000033717.2
ENSMUST00000112115.1 |
Hccs
|
holocytochrome c synthetase |
| chr7_+_51879041 | 3.38 |
ENSMUST00000107591.2
|
Gas2
|
growth arrest specific 2 |
| chr11_-_51756378 | 3.32 |
ENSMUST00000109092.1
ENSMUST00000064297.4 ENSMUST00000109097.2 |
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr19_+_8920358 | 3.25 |
ENSMUST00000096243.5
|
B3gat3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr12_+_80644212 | 3.25 |
ENSMUST00000085245.5
|
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr18_-_20896078 | 3.22 |
ENSMUST00000025177.6
ENSMUST00000097658.1 |
Trappc8
|
trafficking protein particle complex 8 |
| chr8_+_86624043 | 3.22 |
ENSMUST00000034141.9
ENSMUST00000122188.1 |
Lonp2
|
lon peptidase 2, peroxisomal |
| chr2_+_15055274 | 3.21 |
ENSMUST00000069870.3
|
Arl5b
|
ADP-ribosylation factor-like 5B |
| chr1_+_167308649 | 3.21 |
ENSMUST00000097473.4
|
Tmco1
|
transmembrane and coiled-coil domains 1 |
| chr5_-_3803081 | 3.20 |
ENSMUST00000043551.6
|
Ankib1
|
ankyrin repeat and IBR domain containing 1 |
| chrX_-_38564519 | 3.20 |
ENSMUST00000016681.8
|
Cul4b
|
cullin 4B |
| chr9_-_79793378 | 3.17 |
ENSMUST00000034878.5
|
Tmem30a
|
transmembrane protein 30A |
| chr9_-_79793507 | 3.17 |
ENSMUST00000120690.1
|
Tmem30a
|
transmembrane protein 30A |
| chr18_-_56572888 | 3.14 |
ENSMUST00000174518.1
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr16_-_38522662 | 3.13 |
ENSMUST00000002925.5
|
Timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr4_-_149485157 | 3.10 |
ENSMUST00000126896.1
ENSMUST00000105693.1 ENSMUST00000030845.6 |
Nmnat1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr9_-_105495037 | 3.08 |
ENSMUST00000176190.1
ENSMUST00000163879.2 ENSMUST00000112558.2 ENSMUST00000176390.1 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr1_-_36244245 | 3.08 |
ENSMUST00000046875.7
|
Uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr11_-_4160286 | 3.07 |
ENSMUST00000093381.4
ENSMUST00000101626.2 |
Ccdc157
|
coiled-coil domain containing 157 |
| chr12_+_64965742 | 3.05 |
ENSMUST00000066296.7
|
Fam179b
|
family with sequence similarity 179, member B |
| chr15_+_99392882 | 3.03 |
ENSMUST00000023749.8
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chrX_-_12762069 | 3.00 |
ENSMUST00000096495.4
ENSMUST00000076016.5 |
Med14
|
mediator complex subunit 14 |
| chr6_+_8259327 | 2.97 |
ENSMUST00000159378.1
|
Gm16039
|
predicted gene 16039 |
| chr2_-_18392736 | 2.96 |
ENSMUST00000091418.5
ENSMUST00000166495.1 |
Dnajc1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr5_+_3803160 | 2.95 |
ENSMUST00000171023.1
ENSMUST00000080085.4 |
Krit1
|
KRIT1, ankyrin repeat containing |
| chr3_-_84582476 | 2.93 |
ENSMUST00000107687.2
ENSMUST00000098990.3 |
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr1_-_171294937 | 2.92 |
ENSMUST00000111302.3
ENSMUST00000080001.2 |
Ufc1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chr10_+_59221945 | 2.90 |
ENSMUST00000182161.1
|
Sowahc
|
sosondowah ankyrin repeat domain family member C |
| chr19_-_59076069 | 2.89 |
ENSMUST00000047511.7
ENSMUST00000163821.1 |
4930506M07Rik
|
RIKEN cDNA 4930506M07 gene |
| chr9_-_29411736 | 2.89 |
ENSMUST00000115236.1
|
Ntm
|
neurotrimin |
| chr4_-_124850670 | 2.89 |
ENSMUST00000163946.1
ENSMUST00000106190.3 |
1110065P20Rik
|
RIKEN cDNA 1110065P20 gene |
| chr11_+_67052641 | 2.86 |
ENSMUST00000108690.3
ENSMUST00000092996.4 |
Sco1
|
SCO cytochrome oxidase deficient homolog 1 (yeast) |
| chr15_+_99392948 | 2.82 |
ENSMUST00000161250.1
ENSMUST00000160635.1 ENSMUST00000161778.1 |
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr15_+_99393219 | 2.79 |
ENSMUST00000159209.1
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr8_+_94037198 | 2.77 |
ENSMUST00000109556.2
ENSMUST00000093301.2 ENSMUST00000060632.7 |
Ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr7_-_140882274 | 2.75 |
ENSMUST00000026559.7
|
Sirt3
|
sirtuin 3 |
| chr17_-_6948283 | 2.74 |
ENSMUST00000024572.9
|
Rsph3b
|
radial spoke 3B homolog (Chlamydomonas) |
| chr3_+_108571699 | 2.72 |
ENSMUST00000143054.1
|
Taf13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr13_-_90089060 | 2.69 |
ENSMUST00000161396.1
|
Xrcc4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr19_+_6046576 | 2.64 |
ENSMUST00000138532.1
ENSMUST00000129081.1 ENSMUST00000156550.1 |
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr6_+_8259379 | 2.63 |
ENSMUST00000162034.1
ENSMUST00000160705.1 ENSMUST00000159433.1 |
Gm16039
|
predicted gene 16039 |
| chr13_-_59823072 | 2.63 |
ENSMUST00000071703.4
|
Zcchc6
|
zinc finger, CCHC domain containing 6 |
| chr7_+_119895836 | 2.62 |
ENSMUST00000106518.1
ENSMUST00000054440.3 |
Lyrm1
|
LYR motif containing 1 |
| chr10_-_83648713 | 2.62 |
ENSMUST00000020500.7
|
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr4_+_62360695 | 2.61 |
ENSMUST00000084526.5
|
Slc31a1
|
solute carrier family 31, member 1 |
| chr5_-_25100624 | 2.60 |
ENSMUST00000030784.7
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chrX_+_7722267 | 2.54 |
ENSMUST00000125991.1
ENSMUST00000148624.1 |
Wdr45
|
WD repeat domain 45 |
| chr11_-_120573253 | 2.52 |
ENSMUST00000026122.4
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr15_-_55557748 | 2.51 |
ENSMUST00000172387.1
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr9_+_108290433 | 2.51 |
ENSMUST00000035227.6
|
Nicn1
|
nicolin 1 |
| chr13_+_84222286 | 2.51 |
ENSMUST00000057495.8
|
Tmem161b
|
transmembrane protein 161B |
| chr10_-_89732253 | 2.49 |
ENSMUST00000020109.3
|
Actr6
|
ARP6 actin-related protein 6 |
| chr3_-_84582616 | 2.48 |
ENSMUST00000143514.1
|
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr11_-_67052563 | 2.47 |
ENSMUST00000116363.1
|
Adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese dependent |
| chr15_+_35371498 | 2.46 |
ENSMUST00000048646.7
|
Vps13b
|
vacuolar protein sorting 13B (yeast) |
| chr10_+_75893398 | 2.45 |
ENSMUST00000009236.4
|
Derl3
|
Der1-like domain family, member 3 |
| chr10_-_83648631 | 2.45 |
ENSMUST00000146876.2
ENSMUST00000176294.1 |
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chrX_+_107255878 | 2.43 |
ENSMUST00000101294.2
ENSMUST00000118820.1 ENSMUST00000120971.1 |
Gpr174
|
G protein-coupled receptor 174 |
| chr2_+_31572651 | 2.43 |
ENSMUST00000113482.1
|
Fubp3
|
far upstream element (FUSE) binding protein 3 |
| chr6_-_124741374 | 2.41 |
ENSMUST00000004389.5
|
Grcc10
|
gene rich cluster, C10 gene |
| chr7_-_119895446 | 2.41 |
ENSMUST00000098080.2
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr2_-_156144138 | 2.36 |
ENSMUST00000109600.1
ENSMUST00000029147.9 |
Nfs1
|
nitrogen fixation gene 1 (S. cerevisiae) |
| chr19_+_6047081 | 2.35 |
ENSMUST00000025723.8
|
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr9_+_45055166 | 2.35 |
ENSMUST00000114664.1
ENSMUST00000093856.3 |
Mpzl3
|
myelin protein zero-like 3 |
| chr1_+_24678536 | 2.34 |
ENSMUST00000095062.3
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr2_-_168230575 | 2.33 |
ENSMUST00000109193.1
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr4_-_149485202 | 2.30 |
ENSMUST00000119921.1
|
Nmnat1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr3_-_108146080 | 2.30 |
ENSMUST00000000001.4
|
Gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting 3 |
| chr10_-_127070254 | 2.30 |
ENSMUST00000060991.4
|
Tspan31
|
tetraspanin 31 |
| chr6_+_82041623 | 2.29 |
ENSMUST00000042974.8
|
Eva1a
|
eva-1 homolog A (C. elegans) |
| chr9_+_45906513 | 2.28 |
ENSMUST00000039059.6
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr2_-_168230353 | 2.28 |
ENSMUST00000154111.1
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr3_-_63964768 | 2.27 |
ENSMUST00000029402.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr9_+_104063376 | 2.25 |
ENSMUST00000120854.1
|
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr3_+_106721672 | 2.23 |
ENSMUST00000098750.2
ENSMUST00000130105.1 |
Lrif1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr3_-_63964659 | 2.22 |
ENSMUST00000161659.1
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chrX_-_73716145 | 2.22 |
ENSMUST00000002091.5
|
Bcap31
|
B cell receptor associated protein 31 |
| chr18_+_36783222 | 2.22 |
ENSMUST00000019287.8
|
Hars2
|
histidyl-tRNA synthetase 2, mitochondrial (putative) |
| chr14_+_20348159 | 2.20 |
ENSMUST00000090503.4
ENSMUST00000090499.5 ENSMUST00000037698.5 ENSMUST00000051915.6 |
Fam149b
|
family with sequence similarity 149, member B |
| chr17_-_47688028 | 2.20 |
ENSMUST00000113301.1
ENSMUST00000113302.3 |
Tomm6
|
translocase of outer mitochondrial membrane 6 homolog (yeast) |
| chr8_+_72219726 | 2.19 |
ENSMUST00000003123.8
|
Fam32a
|
family with sequence similarity 32, member A |
| chr6_-_119388671 | 2.19 |
ENSMUST00000169744.1
|
Adipor2
|
adiponectin receptor 2 |
| chr9_-_110476637 | 2.16 |
ENSMUST00000111934.1
ENSMUST00000068025.6 |
Klhl18
|
kelch-like 18 |
| chr11_-_70646972 | 2.14 |
ENSMUST00000014750.8
|
Slc25a11
|
solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11 |
| chr6_+_125009261 | 2.14 |
ENSMUST00000112427.1
|
Zfp384
|
zinc finger protein 384 |
| chr3_+_106721893 | 2.14 |
ENSMUST00000106736.2
ENSMUST00000154973.1 ENSMUST00000131330.1 ENSMUST00000150513.1 |
Lrif1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr17_-_35979679 | 2.13 |
ENSMUST00000173724.1
ENSMUST00000172900.1 ENSMUST00000174849.1 |
Prr3
|
proline-rich polypeptide 3 |
| chr9_-_105495475 | 2.12 |
ENSMUST00000176036.1
|
Atp2c1
|
ATPase, Ca++-sequestering |
| chr19_+_6047055 | 2.11 |
ENSMUST00000134667.1
|
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr11_-_72795801 | 2.10 |
ENSMUST00000079681.5
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr16_-_56717286 | 2.09 |
ENSMUST00000121554.1
ENSMUST00000128551.1 |
Tfg
|
Trk-fused gene |
| chrX_+_7722214 | 2.08 |
ENSMUST00000043045.2
ENSMUST00000116634.1 ENSMUST00000115689.3 ENSMUST00000131077.1 ENSMUST00000115688.1 ENSMUST00000116633.1 |
Wdr45
|
WD repeat domain 45 |
| chr16_-_56717182 | 2.07 |
ENSMUST00000141404.1
|
Tfg
|
Trk-fused gene |
| chr5_-_92435114 | 2.07 |
ENSMUST00000135112.1
|
Nup54
|
nucleoporin 54 |
| chr11_+_30771726 | 2.06 |
ENSMUST00000041231.7
|
Psme4
|
proteasome (prosome, macropain) activator subunit 4 |
| chr2_-_75938407 | 2.05 |
ENSMUST00000099996.3
|
Ttc30b
|
tetratricopeptide repeat domain 30B |
| chr2_-_160327494 | 2.05 |
ENSMUST00000099127.2
|
Gm826
|
predicted gene 826 |
| chr2_-_130424242 | 2.04 |
ENSMUST00000089581.4
|
Pced1a
|
PC-esterase domain containing 1A |
| chr11_+_100545607 | 2.02 |
ENSMUST00000092684.5
ENSMUST00000006976.7 |
Ttc25
|
tetratricopeptide repeat domain 25 |
| chr12_-_44210061 | 2.02 |
ENSMUST00000015049.3
|
Dnajb9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr13_+_97137937 | 2.02 |
ENSMUST00000042084.6
ENSMUST00000160139.1 ENSMUST00000161639.1 ENSMUST00000161913.1 ENSMUST00000161825.1 ENSMUST00000161929.1 ENSMUST00000022170.7 |
Gfm2
|
G elongation factor, mitochondrial 2 |
| chr9_+_55208925 | 2.02 |
ENSMUST00000034859.8
|
Fbxo22
|
F-box protein 22 |
| chr6_-_113531575 | 2.01 |
ENSMUST00000032425.5
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr7_+_105640522 | 2.01 |
ENSMUST00000106785.1
ENSMUST00000106786.1 ENSMUST00000106780.1 ENSMUST00000106784.1 |
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr17_-_85090204 | 2.01 |
ENSMUST00000072406.3
ENSMUST00000171795.1 |
Prepl
|
prolyl endopeptidase-like |
| chr9_-_59353430 | 2.00 |
ENSMUST00000026265.6
|
Bbs4
|
Bardet-Biedl syndrome 4 (human) |
| chr15_-_77928925 | 1.99 |
ENSMUST00000109748.2
ENSMUST00000109747.2 ENSMUST00000100486.5 ENSMUST00000005487.5 |
Txn2
|
thioredoxin 2 |
| chr7_-_45062393 | 1.99 |
ENSMUST00000129101.1
|
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr19_-_43524462 | 1.99 |
ENSMUST00000026196.7
|
Got1
|
glutamate oxaloacetate transaminase 1, soluble |
| chr19_-_55315980 | 1.98 |
ENSMUST00000076891.5
|
Zdhhc6
|
zinc finger, DHHC domain containing 6 |
| chr1_-_183297008 | 1.98 |
ENSMUST00000057062.5
|
Brox
|
BRO1 domain and CAAX motif containing |
| chr18_+_56432116 | 1.97 |
ENSMUST00000070166.5
|
Gramd3
|
GRAM domain containing 3 |
| chr1_-_164307443 | 1.94 |
ENSMUST00000027866.4
ENSMUST00000120447.1 ENSMUST00000086032.3 |
Blzf1
|
basic leucine zipper nuclear factor 1 |
| chr10_-_62651194 | 1.94 |
ENSMUST00000020270.4
|
Ddx50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr5_+_143181010 | 1.93 |
ENSMUST00000031574.3
|
4933411G11Rik
|
RIKEN cDNA 4933411G11Rik gene |
| chr6_-_72362382 | 1.92 |
ENSMUST00000114095.1
ENSMUST00000069595.6 ENSMUST00000069580.5 |
Rnf181
|
ring finger protein 181 |
| chr3_+_32529532 | 1.92 |
ENSMUST00000147350.1
|
Mfn1
|
mitofusin 1 |
| chr13_+_54621801 | 1.92 |
ENSMUST00000026991.9
ENSMUST00000137413.1 ENSMUST00000135232.1 ENSMUST00000124752.1 |
Faf2
|
Fas associated factor family member 2 |
| chr16_+_78301673 | 1.92 |
ENSMUST00000114229.2
|
Cxadr
|
coxsackie virus and adenovirus receptor |
| chr4_-_40722307 | 1.91 |
ENSMUST00000181475.1
|
Gm6297
|
predicted gene 6297 |
| chr7_+_80261202 | 1.91 |
ENSMUST00000117989.1
|
Ngrn
|
neugrin, neurite outgrowth associated |
| chr8_+_123212857 | 1.91 |
ENSMUST00000060133.6
|
Spata33
|
spermatogenesis associated 33 |
| chr5_-_92435219 | 1.90 |
ENSMUST00000038514.8
|
Nup54
|
nucleoporin 54 |
| chr3_-_5576111 | 1.90 |
ENSMUST00000165309.1
ENSMUST00000164828.1 ENSMUST00000071280.5 |
Pex2
|
peroxisomal biogenesis factor 2 |
| chr12_+_85110833 | 1.89 |
ENSMUST00000053811.8
|
Dlst
|
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
| chr7_+_110018301 | 1.89 |
ENSMUST00000084731.3
|
Ipo7
|
importin 7 |
| chr18_+_30272747 | 1.89 |
ENSMUST00000115812.3
ENSMUST00000115811.1 ENSMUST00000091978.5 |
Pik3c3
|
phosphoinositide-3-kinase, class 3 |
| chr17_-_36951338 | 1.89 |
ENSMUST00000173540.1
|
Ppp1r11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr5_+_147860615 | 1.89 |
ENSMUST00000031654.6
|
Pomp
|
proteasome maturation protein |
| chr11_-_69920581 | 1.88 |
ENSMUST00000108610.1
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr4_-_116555896 | 1.87 |
ENSMUST00000069674.5
ENSMUST00000106478.2 |
Tmem69
|
transmembrane protein 69 |
| chr7_+_30169861 | 1.85 |
ENSMUST00000085668.4
|
Gm5113
|
predicted gene 5113 |
| chr1_-_160212864 | 1.85 |
ENSMUST00000014370.5
|
Cacybp
|
calcyclin binding protein |
| chr7_+_12834743 | 1.85 |
ENSMUST00000004614.8
|
Zfp110
|
zinc finger protein 110 |
| chrX_+_142228699 | 1.84 |
ENSMUST00000112913.1
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr18_+_7869342 | 1.84 |
ENSMUST00000092112.4
ENSMUST00000172018.1 ENSMUST00000168446.1 |
Wac
|
WW domain containing adaptor with coiled-coil |
| chr1_-_172082757 | 1.83 |
ENSMUST00000003550.4
|
Ncstn
|
nicastrin |
| chr7_+_105640448 | 1.83 |
ENSMUST00000058333.3
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chrX_+_42068398 | 1.82 |
ENSMUST00000115095.2
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr16_-_3872378 | 1.80 |
ENSMUST00000090522.4
|
Zfp597
|
zinc finger protein 597 |
| chr16_-_56717446 | 1.80 |
ENSMUST00000065515.7
|
Tfg
|
Trk-fused gene |
| chr3_+_89418443 | 1.79 |
ENSMUST00000039110.5
ENSMUST00000125036.1 ENSMUST00000154791.1 ENSMUST00000128238.1 ENSMUST00000107417.2 |
Shc1
|
src homology 2 domain-containing transforming protein C1 |
| chr10_-_39899238 | 1.79 |
ENSMUST00000178563.1
|
AA474331
|
expressed sequence AA474331 |
| chr7_+_44896125 | 1.79 |
ENSMUST00000166552.1
ENSMUST00000168207.1 |
Fuz
|
fuzzy homolog (Drosophila) |
| chr3_-_5576233 | 1.78 |
ENSMUST00000059021.4
|
Pex2
|
peroxisomal biogenesis factor 2 |
| chr1_+_16688405 | 1.78 |
ENSMUST00000026881.4
|
Ly96
|
lymphocyte antigen 96 |
| chr16_+_57121705 | 1.78 |
ENSMUST00000166897.1
|
Tomm70a
|
translocase of outer mitochondrial membrane 70 homolog A (yeast) |
| chr4_+_140961203 | 1.78 |
ENSMUST00000010007.8
|
Sdhb
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr2_-_104028287 | 1.78 |
ENSMUST00000056170.3
|
4931422A03Rik
|
RIKEN cDNA 4931422A03 gene |
| chr15_-_100551959 | 1.78 |
ENSMUST00000009877.6
|
Tfcp2
|
transcription factor CP2 |
| chr7_+_46796088 | 1.77 |
ENSMUST00000006774.4
ENSMUST00000165031.1 |
Gtf2h1
|
general transcription factor II H, polypeptide 1 |
| chr2_-_69885557 | 1.76 |
ENSMUST00000060447.6
|
Mettl5
|
methyltransferase like 5 |
| chr5_+_122391878 | 1.75 |
ENSMUST00000102525.4
|
Arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr11_-_82764303 | 1.74 |
ENSMUST00000021040.3
ENSMUST00000100722.4 |
Cct6b
|
chaperonin containing Tcp1, subunit 6b (zeta) |
| chr17_-_86145139 | 1.74 |
ENSMUST00000095187.3
|
Srbd1
|
S1 RNA binding domain 1 |
| chr9_+_55209190 | 1.74 |
ENSMUST00000146201.1
|
Fbxo22
|
F-box protein 22 |
| chr2_+_120977017 | 1.73 |
ENSMUST00000067582.7
|
Tmem62
|
transmembrane protein 62 |
| chr8_+_105326354 | 1.73 |
ENSMUST00000015000.5
ENSMUST00000098453.2 |
Tmem208
|
transmembrane protein 208 |
| chr5_-_143180721 | 1.73 |
ENSMUST00000164068.1
ENSMUST00000049861.4 ENSMUST00000165318.1 |
Rbak
|
RB-associated KRAB repressor |
| chr5_-_108434373 | 1.72 |
ENSMUST00000049628.9
ENSMUST00000118632.1 |
Atp5k
|
ATP synthase, H+ transporting, mitochondrial F1F0 complex, subunit e |
| chrX_+_142228177 | 1.72 |
ENSMUST00000112914.1
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr11_+_29526423 | 1.71 |
ENSMUST00000136351.1
ENSMUST00000020749.6 ENSMUST00000144321.1 ENSMUST00000093239.4 |
Mtif2
|
mitochondrial translational initiation factor 2 |
| chr14_+_65837302 | 1.70 |
ENSMUST00000022614.5
|
Ccdc25
|
coiled-coil domain containing 25 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Etv3_Erf_Fev_Elk4_Elk1_Elk3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 47.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 2.2 | 8.6 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 1.8 | 5.3 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 1.7 | 8.6 | GO:0031438 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 1.5 | 13.7 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 1.4 | 4.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 1.2 | 3.6 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 1.1 | 6.8 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 1.1 | 5.4 | GO:0034627 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 1.1 | 4.3 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 1.1 | 3.2 | GO:2000314 | negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 1.0 | 6.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.9 | 3.8 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.9 | 8.4 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.9 | 2.8 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.9 | 3.6 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.9 | 4.4 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.9 | 2.6 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.8 | 2.4 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.8 | 4.0 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.8 | 2.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.8 | 2.3 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.8 | 2.3 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.7 | 1.5 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.7 | 2.9 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.7 | 2.1 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.7 | 6.8 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.7 | 1.3 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.7 | 2.0 | GO:0006532 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.7 | 4.6 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.6 | 1.9 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.6 | 3.7 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.6 | 2.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.6 | 5.5 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.6 | 2.4 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.6 | 2.4 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.6 | 2.4 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.6 | 2.4 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.6 | 1.8 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.6 | 3.5 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.6 | 1.7 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.5 | 2.7 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.5 | 2.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.5 | 5.2 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.5 | 2.6 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.5 | 1.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.5 | 2.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.5 | 0.5 | GO:0070861 | regulation of protein exit from endoplasmic reticulum(GO:0070861) |
| 0.5 | 1.5 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.5 | 1.5 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.5 | 1.5 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.5 | 10.9 | GO:0072663 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.5 | 2.4 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.5 | 1.9 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.5 | 2.4 | GO:0060296 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.5 | 1.4 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.5 | 3.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.5 | 12.8 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.5 | 7.7 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.4 | 2.7 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.4 | 2.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.4 | 1.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.4 | 1.7 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.4 | 3.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.4 | 3.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.4 | 1.7 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.4 | 1.7 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.4 | 0.8 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.4 | 1.2 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.4 | 1.2 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.4 | 0.8 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.4 | 7.6 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.4 | 1.2 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.4 | 2.7 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.4 | 1.6 | GO:0006203 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.4 | 0.8 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.4 | 2.7 | GO:0006266 | DNA ligation(GO:0006266) |
| 0.4 | 0.8 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.4 | 1.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.4 | 0.7 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.3 | 6.5 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.3 | 1.7 | GO:0021764 | amygdala development(GO:0021764) |
| 0.3 | 4.0 | GO:0006983 | ER overload response(GO:0006983) |
| 0.3 | 2.3 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.3 | 3.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.3 | 2.3 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.3 | 1.0 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.3 | 1.9 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.3 | 3.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.3 | 3.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.3 | 2.3 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.3 | 2.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.3 | 2.2 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.3 | 1.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.3 | 3.5 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.3 | 0.3 | GO:1904809 | dense core granule localization(GO:0032253) dense core granule transport(GO:1901950) regulation of dense core granule transport(GO:1904809) positive regulation of dense core granule transport(GO:1904811) |
| 0.3 | 0.9 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.3 | 0.9 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.3 | 0.9 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.3 | 1.2 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.3 | 6.2 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.3 | 0.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.3 | 1.2 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.3 | 1.8 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.3 | 2.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.3 | 1.5 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.3 | 4.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.3 | 1.1 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.3 | 1.4 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.3 | 4.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.3 | 1.9 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.3 | 2.7 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.3 | 1.4 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.3 | 1.6 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.3 | 1.9 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.3 | 3.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.3 | 1.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.3 | 1.8 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.3 | 4.1 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.3 | 0.8 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.2 | 0.5 | GO:2000152 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) regulation of ubiquitin-specific protease activity(GO:2000152) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.2 | 0.7 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.2 | 3.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 0.7 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 0.7 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.2 | 0.9 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 0.7 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.2 | 1.6 | GO:0090148 | membrane fission(GO:0090148) |
| 0.2 | 1.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.2 | 1.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 1.1 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.2 | 1.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 1.6 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 0.7 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.2 | 0.7 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.2 | 3.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 1.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.2 | 3.0 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 0.4 | GO:0009146 | purine nucleoside triphosphate catabolic process(GO:0009146) |
| 0.2 | 1.7 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.2 | 0.8 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.2 | 1.5 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.2 | 0.8 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 | 0.8 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 | 0.8 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 0.6 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.2 | 0.8 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 0.6 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.2 | 0.8 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.2 | 1.5 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.2 | 1.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 | 8.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.2 | 2.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.2 | 0.9 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 1.1 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.2 | 3.7 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.2 | 0.7 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.2 | 0.5 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.2 | 0.7 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.2 | 1.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.2 | 1.5 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.2 | 3.1 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.2 | 3.6 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.2 | 1.8 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 0.5 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.2 | 0.7 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.2 | 1.6 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.2 | 1.4 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.2 | 1.7 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.2 | 2.3 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 | 3.0 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 2.7 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.2 | 4.1 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 7.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 1.0 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 1.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 2.2 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.2 | 3.6 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.2 | 0.6 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.2 | 1.3 | GO:0090043 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.2 | 0.8 | GO:0072386 | stress granule disassembly(GO:0035617) plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 1.6 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.2 | 0.5 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 2.2 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.2 | 0.9 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 0.6 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 1.9 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 1.0 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 4.3 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.1 | 1.0 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 1.4 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.5 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.5 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.4 | GO:0097466 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.1 | 8.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.4 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 4.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.5 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.1 | 1.0 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 1.6 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 1.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.4 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.1 | 0.6 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.1 | 2.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.2 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 2.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.3 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.1 | 0.7 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.6 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.1 | 1.9 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.7 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.1 | 5.7 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.1 | 0.9 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 0.4 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.1 | GO:0046619 | optic placode formation(GO:0001743) optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 1.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.3 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.1 | 0.6 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.3 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 1.6 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 2.0 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.1 | 0.3 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.3 | GO:0072422 | signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
| 0.1 | 0.3 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.1 | 0.1 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.1 | 0.6 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.1 | 0.3 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 3.0 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.1 | 0.2 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 0.2 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 0.8 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 0.3 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 1.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 2.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.6 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 1.0 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.8 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.3 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.5 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.1 | 1.6 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 1.9 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.1 | 1.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.4 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 0.3 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.1 | 0.6 | GO:1902741 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.1 | 1.5 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 2.7 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 0.7 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 6.0 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 2.0 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 1.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.7 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.5 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 1.8 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 1.3 | GO:0045019 | negative regulation of nitric oxide biosynthetic process(GO:0045019) negative regulation of nitric oxide metabolic process(GO:1904406) |
| 0.1 | 1.9 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.1 | 0.4 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.6 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.7 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 1.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.6 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.1 | 0.5 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 0.5 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.3 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 1.7 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 1.1 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.5 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 0.4 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.3 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 2.6 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 0.6 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.6 | GO:1903798 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.1 | 0.5 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 0.4 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 0.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.3 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.1 | 1.2 | GO:0003283 | atrial septum development(GO:0003283) |
| 0.1 | 0.3 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 0.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 0.6 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.1 | 0.5 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.1 | 1.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 2.0 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 0.8 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 2.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 0.1 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.1 | 4.7 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 0.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.1 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.1 | 2.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.2 | GO:0021995 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 0.3 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 0.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 1.1 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.1 | 1.1 | GO:1903540 | neurotransmitter receptor transport to plasma membrane(GO:0098877) neurotransmitter receptor transport to postsynaptic membrane(GO:0098969) establishment of protein localization to postsynaptic membrane(GO:1903540) |
| 0.1 | 1.0 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.5 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.6 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 0.3 | GO:0021756 | striatum development(GO:0021756) |
| 0.1 | 1.4 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.1 | 0.5 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 1.9 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 0.7 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.1 | 0.3 | GO:1902861 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) copper ion import across plasma membrane(GO:0098705) copper ion import into cell(GO:1902861) |
| 0.1 | 0.1 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.1 | 0.7 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.1 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.1 | 0.2 | GO:1903537 | meiotic sister chromatid cohesion, centromeric(GO:0051754) meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 1.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 6.0 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 0.3 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.2 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.1 | 0.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.9 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.2 | GO:1990401 | embryonic lung development(GO:1990401) |
| 0.1 | 2.2 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 0.6 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 0.2 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 0.5 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.5 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 1.7 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 1.8 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 1.2 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 1.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.9 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 0.1 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 0.2 | GO:0046294 | ethanol oxidation(GO:0006069) formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.3 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.3 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 0.0 | 0.2 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.2 | GO:0032667 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.0 | 1.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.8 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.0 | 0.1 | GO:1902276 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.0 | 0.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.3 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.2 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.2 | GO:2000303 | regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.1 | GO:2000065 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) regulation of cortisol biosynthetic process(GO:2000064) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 1.0 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.8 | GO:0090311 | regulation of protein deacetylation(GO:0090311) |
| 0.0 | 0.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.3 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.9 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.3 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.5 | GO:0006206 | pyrimidine nucleobase metabolic process(GO:0006206) |
| 0.0 | 0.5 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.7 | GO:0044253 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 | 0.2 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.0 | 0.4 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 | 1.1 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.9 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 1.6 | GO:0035904 | aorta development(GO:0035904) |
| 0.0 | 0.5 | GO:0016045 | detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.0 | 1.2 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.5 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.2 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 2.0 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 1.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.0 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.7 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.2 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.2 | GO:0003179 | heart valve morphogenesis(GO:0003179) |
| 0.0 | 0.6 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.1 | GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904953) |
| 0.0 | 1.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.2 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.3 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.5 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 1.5 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.7 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.4 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.8 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 0.2 | GO:0035357 | peroxisome proliferator activated receptor signaling pathway(GO:0035357) |
| 0.0 | 0.5 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 2.5 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.0 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 2.8 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.1 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 3.8 | GO:0030198 | extracellular matrix organization(GO:0030198) |
| 0.0 | 1.0 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 1.0 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.5 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.6 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 1.0 | GO:0042308 | negative regulation of protein import into nucleus(GO:0042308) negative regulation of protein import(GO:1904590) |
| 0.0 | 1.0 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.4 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 1.1 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 1.6 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.1 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.1 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.3 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.1 | GO:0097205 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.0 | 0.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.2 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.4 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.5 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.3 | GO:0019674 | NAD metabolic process(GO:0019674) |
| 0.0 | 0.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 1.5 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0032308 | positive regulation of prostaglandin secretion(GO:0032308) |
| 0.0 | 0.9 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.1 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.1 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.2 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 2.1 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.0 | 0.1 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 0.1 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.2 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.0 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.0 | 0.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.5 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.2 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.0 | 0.2 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.7 | GO:0007584 | response to nutrient(GO:0007584) |
| 0.0 | 0.1 | GO:0021942 | layer formation in cerebral cortex(GO:0021819) radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.2 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.0 | 0.4 | GO:0030858 | positive regulation of epithelial cell differentiation(GO:0030858) |
| 0.0 | 0.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.8 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.3 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 47.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 2.3 | 13.7 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 1.4 | 7.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 1.2 | 4.6 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.8 | 4.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.8 | 3.1 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.7 | 3.0 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.7 | 2.2 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.7 | 2.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.6 | 1.9 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.6 | 2.5 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.6 | 6.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.6 | 2.3 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.5 | 6.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.5 | 2.6 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.5 | 2.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.5 | 3.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.5 | 3.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.4 | 0.4 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.4 | 6.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.4 | 8.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.4 | 2.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.4 | 11.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.4 | 3.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.4 | 1.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.4 | 1.9 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.4 | 2.6 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.4 | 2.9 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.4 | 1.8 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 3.0 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.3 | 1.6 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.3 | 2.3 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.3 | 3.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.3 | 3.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.3 | 3.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.3 | 0.9 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
| 0.3 | 2.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.3 | 6.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.3 | 0.9 | GO:0055087 | Ski complex(GO:0055087) |
| 0.3 | 7.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.3 | 1.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 1.4 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.3 | 2.7 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.3 | 0.8 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.3 | 1.8 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.3 | 1.6 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.3 | 7.0 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 1.0 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.3 | 3.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 2.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 2.4 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 3.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 2.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.2 | 0.7 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.2 | 3.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 3.3 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.2 | 1.9 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 0.9 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.2 | 0.9 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 1.8 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.2 | 1.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 1.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.2 | 1.0 | GO:0036396 | MIS complex(GO:0036396) |
| 0.2 | 2.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.2 | 0.6 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 1.8 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.2 | 2.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 1.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 2.5 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) |
| 0.2 | 3.9 | GO:0038201 | TOR complex(GO:0038201) |
| 0.2 | 1.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 1.7 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.2 | 2.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 8.2 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 8.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 0.5 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 1.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.2 | 1.0 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 2.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 7.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 8.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 5.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.4 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 2.6 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 1.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 4.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.6 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 0.4 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 1.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.6 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 3.0 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 5.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.5 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.8 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 0.3 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.5 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 5.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 4.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.7 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.1 | 10.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.9 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 2.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 2.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 5.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 4.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.4 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) |
| 0.1 | 0.4 | GO:0034448 | EGO complex(GO:0034448) |
| 0.1 | 1.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.6 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.1 | 0.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 1.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 2.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.0 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 3.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 1.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.2 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.1 | 16.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 0.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 10.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 28.9 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.1 | 0.2 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 0.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.4 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 2.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 3.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.6 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 0.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.4 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 4.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.9 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 4.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.8 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.0 | 1.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.4 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0005740 | mitochondrial envelope(GO:0005740) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.5 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 1.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 4.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.7 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 3.2 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.4 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 0.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 22.5 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 3.3 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.2 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.6 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 1.8 | 10.7 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 1.4 | 8.6 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 1.2 | 7.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 1.2 | 3.7 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 1.2 | 4.6 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 1.1 | 5.4 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 1.1 | 4.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 1.0 | 6.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 1.0 | 2.9 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.9 | 2.7 | GO:0033699 | DNA 5'-adenosine monophosphate hydrolase activity(GO:0033699) |
| 0.8 | 7.6 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.8 | 2.5 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.8 | 3.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.8 | 4.7 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.8 | 3.1 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.8 | 2.4 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.8 | 2.3 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.8 | 2.3 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.8 | 2.3 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.7 | 1.5 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.7 | 2.9 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.7 | 2.7 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.7 | 2.0 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.7 | 2.0 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.6 | 3.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.6 | 2.6 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.6 | 1.9 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.6 | 1.7 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.5 | 2.6 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.5 | 3.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.5 | 2.5 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.5 | 1.5 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.5 | 1.4 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.5 | 2.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.5 | 2.3 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.4 | 1.8 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.4 | 3.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.4 | 1.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.4 | 1.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.4 | 1.7 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.4 | 1.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.4 | 1.2 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.4 | 1.6 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.4 | 3.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.4 | 2.7 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.4 | 4.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.4 | 1.2 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.4 | 6.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.4 | 3.6 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.4 | 1.8 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.3 | 3.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.3 | 2.4 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.3 | 3.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.3 | 2.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 10.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.3 | 0.9 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.3 | 1.6 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.3 | 2.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.3 | 1.8 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.3 | 1.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.3 | 2.4 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 2.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.3 | 2.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.3 | 1.7 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.3 | 0.8 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.3 | 2.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.3 | 0.8 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.3 | 1.0 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.3 | 2.6 | GO:0035473 | lipase binding(GO:0035473) |
| 0.2 | 0.7 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.2 | 0.5 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.2 | 0.7 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 3.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 1.9 | GO:0002046 | opsin binding(GO:0002046) |
| 0.2 | 0.7 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 3.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 2.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 0.9 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.2 | 1.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 9.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 2.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 0.7 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.2 | 2.7 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.2 | 1.7 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 1.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 0.6 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.2 | 6.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 1.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 0.8 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.2 | 0.8 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 0.4 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.2 | 3.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 0.6 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.2 | 0.8 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.2 | 1.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.2 | 0.6 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 0.7 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.2 | 0.7 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.2 | 0.2 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.2 | 1.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.2 | 0.5 | GO:0052744 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.2 | 0.9 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.2 | 2.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 1.9 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.2 | 2.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 0.3 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 1.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 1.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 0.5 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.2 | 1.0 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.2 | 1.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 0.3 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.2 | 0.5 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.2 | 1.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 1.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.2 | 1.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 1.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.2 | 1.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 1.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 1.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.2 | 0.6 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 1.6 | GO:0046977 | TAP binding(GO:0046977) |
| 0.1 | 1.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.4 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.1 | 1.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.6 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 7.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 1.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 2.0 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 0.5 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.9 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 0.9 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 2.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 3.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 5.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 3.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 2.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 2.9 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.8 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.6 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 3.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 3.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.6 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 1.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 1.0 | GO:0098847 | sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 0.6 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.8 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 0.8 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 2.4 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.1 | 0.7 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 3.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 1.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 2.0 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 1.8 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.1 | 1.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.2 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.6 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.1 | 0.9 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 1.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 4.1 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 0.9 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.1 | 0.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 1.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 2.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 2.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.6 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.1 | 1.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 0.5 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.3 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 8.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 0.9 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 2.1 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.4 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 19.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.1 | 1.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.5 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.1 | 1.4 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 3.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.1 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.3 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 2.1 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 1.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.7 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 1.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.4 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 0.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 1.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 3.7 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.1 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.5 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 1.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.2 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.1 | 0.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 1.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.5 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 1.0 | GO:0046030 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.1 | 1.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.1 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.2 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.1 | 1.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.6 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.8 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.2 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.5 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 3.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.2 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.1 | 1.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 2.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.3 | GO:1904315 | GABA-gated chloride ion channel activity(GO:0022851) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.5 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 0.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 3.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 11.1 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.1 | 4.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 1.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 1.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.2 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.4 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 1.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.9 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.7 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 12.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 1.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.3 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 1.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.2 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.0 | GO:0001083 | transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 1.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 4.0 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 23.1 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.5 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 3.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.5 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 3.6 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.2 | GO:0016505 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 1.9 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 5.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 2.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 1.5 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.5 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 2.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 2.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.0 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.2 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 0.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 2.1 | GO:0022832 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.0 | 0.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 2.2 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 1.1 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.2 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.0 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.0 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 21.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 15.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 6.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 4.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 1.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 2.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 5.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 7.2 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 3.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 2.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 0.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 0.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 3.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 2.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 2.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.8 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.3 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.2 | PID INSULIN PATHWAY | Insulin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.6 | 9.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.4 | 5.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.3 | 6.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.3 | 4.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 6.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 1.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 3.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.2 | 2.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 1.5 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.2 | 2.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 2.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 8.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 10.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 9.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 2.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 3.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 4.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 2.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 0.4 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 2.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 5.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 6.3 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 2.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 4.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 3.5 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 3.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 3.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 1.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.4 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 2.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 0.3 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.9 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 3.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.5 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 3.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 7.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 3.8 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 1.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.1 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 3.0 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.3 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 1.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 2.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |