Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Esrrb_Esrra
Z-value: 0.94
Transcription factors associated with Esrrb_Esrra
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
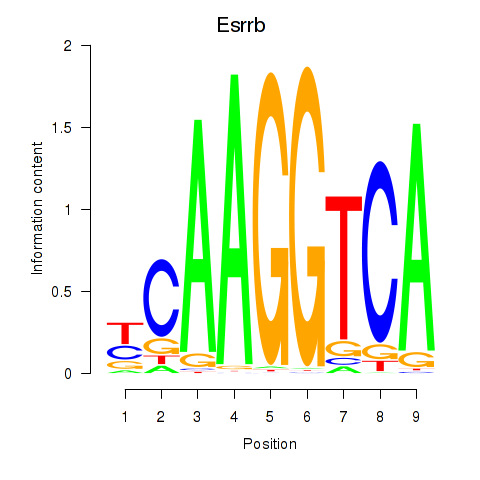
Esrrb
|
ENSMUSG00000021255.11 | estrogen related receptor, beta |
|
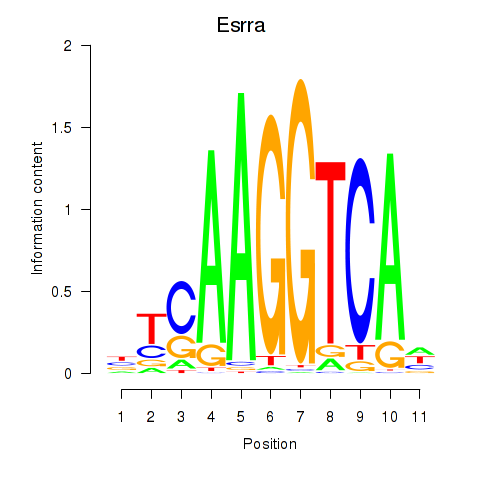
Esrra
|
ENSMUSG00000024955.7 | estrogen related receptor, alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Esrra | mm10_v2_chr19_-_6921804_6921834 | 0.52 | 1.1e-03 | Click! |
| Esrrb | mm10_v2_chr12_+_86361112_86361131 | 0.17 | 3.3e-01 | Click! |
Activity profile of Esrrb_Esrra motif
Sorted Z-values of Esrrb_Esrra motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_43524462 | 2.85 |
ENSMUST00000026196.7
|
Got1
|
glutamate oxaloacetate transaminase 1, soluble |
| chr8_-_104624266 | 1.92 |
ENSMUST00000163783.2
|
Cdh16
|
cadherin 16 |
| chr4_+_34893772 | 1.86 |
ENSMUST00000029975.3
ENSMUST00000135871.1 ENSMUST00000108130.1 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr7_-_141429351 | 1.77 |
ENSMUST00000164387.1
ENSMUST00000137488.1 ENSMUST00000084436.3 |
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr2_-_28563362 | 1.71 |
ENSMUST00000028161.5
|
Cel
|
carboxyl ester lipase |
| chr3_-_20275659 | 1.58 |
ENSMUST00000011607.5
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chrX_-_8018492 | 1.49 |
ENSMUST00000033503.2
|
Glod5
|
glyoxalase domain containing 5 |
| chr11_+_3989924 | 1.43 |
ENSMUST00000109981.1
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr7_-_142576492 | 1.37 |
ENSMUST00000140716.1
|
H19
|
H19 fetal liver mRNA |
| chr1_+_72824482 | 1.36 |
ENSMUST00000047328.4
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr4_-_106492283 | 1.28 |
ENSMUST00000054472.3
|
Bsnd
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr9_-_121792478 | 1.23 |
ENSMUST00000035110.4
|
Hhatl
|
hedgehog acyltransferase-like |
| chrX_-_162964557 | 1.21 |
ENSMUST00000038769.2
|
S100g
|
S100 calcium binding protein G |
| chr4_-_63662910 | 1.05 |
ENSMUST00000184252.1
|
Gm11214
|
predicted gene 11214 |
| chr11_+_115462464 | 1.00 |
ENSMUST00000106532.3
ENSMUST00000092445.5 ENSMUST00000153466.1 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr7_-_141429433 | 0.99 |
ENSMUST00000124444.1
|
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr15_-_79285470 | 0.99 |
ENSMUST00000170955.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr15_+_85510812 | 0.95 |
ENSMUST00000079690.2
|
Gm4825
|
predicted pseudogene 4825 |
| chr4_+_129906205 | 0.94 |
ENSMUST00000145196.1
ENSMUST00000141731.1 ENSMUST00000129149.1 |
E330017L17Rik
|
RIKEN cDNA E330017L17 gene |
| chr4_-_128962420 | 0.93 |
ENSMUST00000119354.1
ENSMUST00000106068.1 ENSMUST00000030581.3 |
Adc
|
arginine decarboxylase |
| chr15_+_35296090 | 0.88 |
ENSMUST00000022952.4
|
Osr2
|
odd-skipped related 2 |
| chr16_-_4789984 | 0.87 |
ENSMUST00000004173.5
|
Cdip1
|
cell death inducing Trp53 target 1 |
| chr2_+_121358591 | 0.86 |
ENSMUST00000000317.6
ENSMUST00000129130.1 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr16_-_4789887 | 0.85 |
ENSMUST00000117713.1
|
Cdip1
|
cell death inducing Trp53 target 1 |
| chr10_+_80264942 | 0.85 |
ENSMUST00000105362.1
ENSMUST00000105361.3 |
Dazap1
|
DAZ associated protein 1 |
| chr2_+_121357714 | 0.83 |
ENSMUST00000125812.1
ENSMUST00000078222.2 ENSMUST00000125221.1 ENSMUST00000150271.1 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr17_-_56117577 | 0.80 |
ENSMUST00000019808.5
|
Plin5
|
perilipin 5 |
| chr6_+_90619241 | 0.77 |
ENSMUST00000032177.8
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr14_+_32321987 | 0.77 |
ENSMUST00000022480.7
|
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr17_-_56117265 | 0.73 |
ENSMUST00000113072.2
|
Plin5
|
perilipin 5 |
| chr1_+_134193432 | 0.71 |
ENSMUST00000038445.6
|
Mybph
|
myosin binding protein H |
| chr15_+_102966794 | 0.71 |
ENSMUST00000001699.7
|
Hoxc10
|
homeobox C10 |
| chr11_+_121702591 | 0.71 |
ENSMUST00000125580.1
|
Metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr4_+_103114712 | 0.70 |
ENSMUST00000143417.1
|
Mier1
|
mesoderm induction early response 1 homolog (Xenopus laevis |
| chr6_-_113501818 | 0.70 |
ENSMUST00000101059.1
|
Prrt3
|
proline-rich transmembrane protein 3 |
| chr2_-_73911323 | 0.70 |
ENSMUST00000111996.1
ENSMUST00000018914.2 |
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) |
| chr9_-_96437434 | 0.68 |
ENSMUST00000070500.2
|
BC043934
|
cDNA sequence BC043934 |
| chr8_-_105471481 | 0.67 |
ENSMUST00000014990.6
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr11_+_5058111 | 0.66 |
ENSMUST00000037218.1
|
Rasl10a
|
RAS-like, family 10, member A |
| chr10_-_68541842 | 0.65 |
ENSMUST00000020103.2
|
1700040L02Rik
|
RIKEN cDNA 1700040L02 gene |
| chr11_-_4704334 | 0.63 |
ENSMUST00000058407.5
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr2_+_32628390 | 0.63 |
ENSMUST00000156578.1
|
Ak1
|
adenylate kinase 1 |
| chr11_+_7063423 | 0.63 |
ENSMUST00000020706.4
|
Adcy1
|
adenylate cyclase 1 |
| chr7_-_19715395 | 0.63 |
ENSMUST00000032555.9
ENSMUST00000093552.5 |
Tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr2_-_30415389 | 0.62 |
ENSMUST00000142096.1
|
Crat
|
carnitine acetyltransferase |
| chr6_+_78425973 | 0.61 |
ENSMUST00000079926.5
|
Reg1
|
regenerating islet-derived 1 |
| chr5_-_147322435 | 0.61 |
ENSMUST00000100433.4
|
Urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5) decarboxylase |
| chr15_-_79285502 | 0.60 |
ENSMUST00000165408.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr2_-_30415767 | 0.60 |
ENSMUST00000102855.1
ENSMUST00000028207.6 |
Crat
|
carnitine acetyltransferase |
| chr17_-_25868727 | 0.59 |
ENSMUST00000026828.5
|
Fam195a
|
family with sequence similarity 195, member A |
| chr16_+_97356721 | 0.59 |
ENSMUST00000047275.6
|
Bace2
|
beta-site APP-cleaving enzyme 2 |
| chr10_+_80265035 | 0.58 |
ENSMUST00000092305.5
|
Dazap1
|
DAZ associated protein 1 |
| chr5_+_118027743 | 0.58 |
ENSMUST00000031304.7
|
Tesc
|
tescalcin |
| chr5_-_137613759 | 0.57 |
ENSMUST00000155251.1
ENSMUST00000124693.1 |
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr13_+_108214389 | 0.55 |
ENSMUST00000022207.8
|
Elovl7
|
ELOVL family member 7, elongation of long chain fatty acids (yeast) |
| chr18_+_77773956 | 0.54 |
ENSMUST00000114748.1
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 |
| chr17_-_25880236 | 0.54 |
ENSMUST00000176696.1
ENSMUST00000095487.5 |
Wfikkn1
|
WAP, FS, Ig, KU, and NTR-containing protein 1 |
| chr10_+_128194446 | 0.54 |
ENSMUST00000044776.6
|
Gls2
|
glutaminase 2 (liver, mitochondrial) |
| chr7_-_4445637 | 0.53 |
ENSMUST00000008579.7
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr1_-_134235420 | 0.53 |
ENSMUST00000038191.6
ENSMUST00000086465.4 |
Adora1
|
adenosine A1 receptor |
| chr14_-_30915387 | 0.53 |
ENSMUST00000166622.1
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chrX_+_135993820 | 0.53 |
ENSMUST00000058119.7
|
Arxes2
|
adipocyte-related X-chromosome expressed sequence 2 |
| chr1_+_75546258 | 0.53 |
ENSMUST00000124341.1
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr16_+_32608920 | 0.52 |
ENSMUST00000023486.8
|
Tfrc
|
transferrin receptor |
| chr1_+_74791516 | 0.52 |
ENSMUST00000006718.8
|
Wnt10a
|
wingless related MMTV integration site 10a |
| chr9_+_65101453 | 0.52 |
ENSMUST00000077696.6
ENSMUST00000035499.4 ENSMUST00000166273.1 |
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr4_-_6275629 | 0.51 |
ENSMUST00000029905.1
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr16_+_22951072 | 0.51 |
ENSMUST00000023590.8
|
Hrg
|
histidine-rich glycoprotein |
| chr19_-_5085483 | 0.50 |
ENSMUST00000140389.1
ENSMUST00000151413.1 ENSMUST00000077066.7 |
Tmem151a
|
transmembrane protein 151A |
| chr6_+_70726430 | 0.50 |
ENSMUST00000103410.1
|
Igkc
|
immunoglobulin kappa constant |
| chr13_-_9878998 | 0.50 |
ENSMUST00000063093.9
|
Chrm3
|
cholinergic receptor, muscarinic 3, cardiac |
| chr15_-_86186136 | 0.50 |
ENSMUST00000044332.9
|
Cerk
|
ceramide kinase |
| chr7_+_28440927 | 0.49 |
ENSMUST00000078845.6
|
Gmfg
|
glia maturation factor, gamma |
| chr3_+_67374091 | 0.49 |
ENSMUST00000077916.5
|
Mlf1
|
myeloid leukemia factor 1 |
| chr4_-_137430517 | 0.48 |
ENSMUST00000102522.4
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr5_-_5694024 | 0.48 |
ENSMUST00000115425.2
ENSMUST00000115427.1 ENSMUST00000115424.2 ENSMUST00000015797.4 |
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr3_-_75270073 | 0.47 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr7_-_4445595 | 0.47 |
ENSMUST00000119485.1
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr7_-_105600103 | 0.47 |
ENSMUST00000033185.8
|
Hpx
|
hemopexin |
| chr14_+_25694170 | 0.47 |
ENSMUST00000022419.6
|
Ppif
|
peptidylprolyl isomerase F (cyclophilin F) |
| chr11_+_96251100 | 0.46 |
ENSMUST00000129907.2
|
Gm53
|
predicted gene 53 |
| chr10_-_71344933 | 0.46 |
ENSMUST00000045887.8
|
Cisd1
|
CDGSH iron sulfur domain 1 |
| chr3_+_67374116 | 0.46 |
ENSMUST00000061322.8
|
Mlf1
|
myeloid leukemia factor 1 |
| chr9_+_106247930 | 0.46 |
ENSMUST00000180701.1
|
4930500F10Rik
|
RIKEN cDNA 4930500F10 gene |
| chr5_+_52363925 | 0.46 |
ENSMUST00000101208.4
|
Sod3
|
superoxide dismutase 3, extracellular |
| chr6_+_113531675 | 0.46 |
ENSMUST00000036340.5
ENSMUST00000101051.2 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr10_+_128194631 | 0.45 |
ENSMUST00000123291.1
|
Gls2
|
glutaminase 2 (liver, mitochondrial) |
| chr12_+_102554966 | 0.44 |
ENSMUST00000021610.5
|
Chga
|
chromogranin A |
| chr7_+_46847128 | 0.44 |
ENSMUST00000005051.4
|
Ldha
|
lactate dehydrogenase A |
| chrX_-_8145713 | 0.43 |
ENSMUST00000115615.2
ENSMUST00000115616.1 ENSMUST00000115621.2 |
Rbm3
|
RNA binding motif protein 3 |
| chr7_-_100547620 | 0.43 |
ENSMUST00000064334.2
|
D630004N19Rik
|
RIKEN cDNA D630004N19 gene |
| chr8_-_84044982 | 0.43 |
ENSMUST00000061923.4
|
Rln3
|
relaxin 3 |
| chrX_-_8145679 | 0.42 |
ENSMUST00000115619.1
ENSMUST00000115617.3 ENSMUST00000040010.3 |
Rbm3
|
RNA binding motif protein 3 |
| chr10_+_115569986 | 0.42 |
ENSMUST00000173620.1
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr4_-_49408042 | 0.42 |
ENSMUST00000081541.2
|
Acnat2
|
acyl-coenzyme A amino acid N-acyltransferase 2 |
| chr7_-_93081027 | 0.41 |
ENSMUST00000098303.1
|
Gm9934
|
predicted gene 9934 |
| chr11_-_69560186 | 0.41 |
ENSMUST00000004036.5
|
Efnb3
|
ephrin B3 |
| chr16_+_26463135 | 0.41 |
ENSMUST00000161053.1
ENSMUST00000115302.1 |
Cldn16
|
claudin 16 |
| chr19_-_3912711 | 0.40 |
ENSMUST00000075092.6
|
Ndufs8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8 |
| chr15_+_100761741 | 0.40 |
ENSMUST00000023776.6
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr5_+_129725063 | 0.39 |
ENSMUST00000086046.3
|
Gbas
|
glioblastoma amplified sequence |
| chr10_-_96409038 | 0.39 |
ENSMUST00000179683.1
|
Gm20091
|
predicted gene, 20091 |
| chr17_-_31144271 | 0.39 |
ENSMUST00000024826.7
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr15_+_88862178 | 0.39 |
ENSMUST00000042818.9
|
Pim3
|
proviral integration site 3 |
| chr16_-_20426375 | 0.39 |
ENSMUST00000079158.6
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr19_+_8591254 | 0.38 |
ENSMUST00000010251.3
ENSMUST00000170817.1 |
Slc22a8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr17_+_47737030 | 0.38 |
ENSMUST00000086932.3
|
Tfeb
|
transcription factor EB |
| chr19_-_11266122 | 0.37 |
ENSMUST00000169159.1
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr14_+_58072686 | 0.37 |
ENSMUST00000022545.7
|
Fgf9
|
fibroblast growth factor 9 |
| chr1_-_84696182 | 0.37 |
ENSMUST00000049126.6
|
Dner
|
delta/notch-like EGF-related receptor |
| chr11_-_53430779 | 0.37 |
ENSMUST00000061326.4
ENSMUST00000109021.3 |
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr7_+_83631959 | 0.36 |
ENSMUST00000075418.7
ENSMUST00000117410.1 |
Stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr4_+_140961203 | 0.36 |
ENSMUST00000010007.8
|
Sdhb
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr11_+_74649462 | 0.36 |
ENSMUST00000092915.5
ENSMUST00000117818.1 |
Cluh
|
clustered mitochondria (cluA/CLU1) homolog |
| chr13_+_55399648 | 0.35 |
ENSMUST00000057167.7
|
Slc34a1
|
solute carrier family 34 (sodium phosphate), member 1 |
| chr1_-_175625580 | 0.35 |
ENSMUST00000027810.7
|
Fh1
|
fumarate hydratase 1 |
| chr4_-_137409777 | 0.35 |
ENSMUST00000024200.6
|
Gm13011
|
predicted gene 13011 |
| chr18_-_15063560 | 0.35 |
ENSMUST00000168989.1
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr17_+_43952999 | 0.35 |
ENSMUST00000177857.1
|
Rcan2
|
regulator of calcineurin 2 |
| chr19_-_47090610 | 0.35 |
ENSMUST00000096014.3
|
Usmg5
|
upregulated during skeletal muscle growth 5 |
| chr13_-_51567084 | 0.35 |
ENSMUST00000021898.5
|
Shc3
|
src homology 2 domain-containing transforming protein C3 |
| chr9_-_50603792 | 0.35 |
ENSMUST00000000175.4
|
Sdhd
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr15_+_76343504 | 0.34 |
ENSMUST00000023210.6
|
Cyc1
|
cytochrome c-1 |
| chr9_+_108080436 | 0.34 |
ENSMUST00000035211.7
ENSMUST00000162886.1 |
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr5_-_123182704 | 0.34 |
ENSMUST00000154713.1
ENSMUST00000031398.7 |
Hpd
|
4-hydroxyphenylpyruvic acid dioxygenase |
| chr9_-_21312255 | 0.34 |
ENSMUST00000115433.3
ENSMUST00000003397.7 |
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr11_+_120484613 | 0.33 |
ENSMUST00000043627.7
|
Mrpl12
|
mitochondrial ribosomal protein L12 |
| chr19_+_6996114 | 0.33 |
ENSMUST00000088223.5
|
Trpt1
|
tRNA phosphotransferase 1 |
| chr11_+_120361506 | 0.33 |
ENSMUST00000026445.2
|
Fscn2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr9_-_106247730 | 0.33 |
ENSMUST00000112524.2
ENSMUST00000074082.6 |
Alas1
|
aminolevulinic acid synthase 1 |
| chr9_-_107668967 | 0.33 |
ENSMUST00000177567.1
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr2_-_54085542 | 0.33 |
ENSMUST00000100089.2
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr8_-_105933832 | 0.33 |
ENSMUST00000034368.6
|
Ctrl
|
chymotrypsin-like |
| chr5_-_5694269 | 0.33 |
ENSMUST00000148333.1
|
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr15_-_79804717 | 0.33 |
ENSMUST00000023057.8
|
Nptxr
|
neuronal pentraxin receptor |
| chr16_-_91931643 | 0.32 |
ENSMUST00000023677.3
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr11_-_33276334 | 0.32 |
ENSMUST00000183831.1
|
Gm12117
|
predicted gene 12117 |
| chr4_+_137681663 | 0.32 |
ENSMUST00000047243.5
|
Rap1gap
|
Rap1 GTPase-activating protein |
| chr9_-_63399216 | 0.32 |
ENSMUST00000168665.1
|
2300009A05Rik
|
RIKEN cDNA 2300009A05 gene |
| chr7_-_66427469 | 0.32 |
ENSMUST00000015278.7
|
Aldh1a3
|
aldehyde dehydrogenase family 1, subfamily A3 |
| chr4_+_138434647 | 0.31 |
ENSMUST00000044058.4
ENSMUST00000105813.1 ENSMUST00000105815.1 |
Mul1
|
mitochondrial ubiquitin ligase activator of NFKB 1 |
| chr8_+_117095854 | 0.31 |
ENSMUST00000034308.8
ENSMUST00000167370.1 ENSMUST00000176860.1 |
Bcmo1
|
beta-carotene 15,15'-monooxygenase |
| chr1_-_172895048 | 0.31 |
ENSMUST00000027824.5
|
Apcs
|
serum amyloid P-component |
| chr2_-_127541385 | 0.31 |
ENSMUST00000103214.2
|
Prom2
|
prominin 2 |
| chr8_-_70523085 | 0.31 |
ENSMUST00000137610.1
ENSMUST00000121623.1 ENSMUST00000093456.5 ENSMUST00000118850.1 |
Kxd1
|
KxDL motif containing 1 |
| chr18_-_79109391 | 0.31 |
ENSMUST00000025430.8
ENSMUST00000161465.2 |
Setbp1
|
SET binding protein 1 |
| chr2_+_91035613 | 0.30 |
ENSMUST00000111445.3
ENSMUST00000111446.3 ENSMUST00000050323.5 |
Rapsn
|
receptor-associated protein of the synapse |
| chr16_+_8637674 | 0.30 |
ENSMUST00000023396.9
|
Pmm2
|
phosphomannomutase 2 |
| chr17_-_32420965 | 0.30 |
ENSMUST00000170392.1
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr11_+_70764209 | 0.30 |
ENSMUST00000060444.5
|
Zfp3
|
zinc finger protein 3 |
| chr1_+_82586942 | 0.30 |
ENSMUST00000113457.2
|
Col4a3
|
collagen, type IV, alpha 3 |
| chr7_+_28441026 | 0.30 |
ENSMUST00000135686.1
|
Gmfg
|
glia maturation factor, gamma |
| chr6_-_115762346 | 0.30 |
ENSMUST00000166254.1
ENSMUST00000170625.1 |
Tmem40
|
transmembrane protein 40 |
| chr4_+_123233556 | 0.29 |
ENSMUST00000040821.4
|
Heyl
|
hairy/enhancer-of-split related with YRPW motif-like |
| chr5_+_33658123 | 0.29 |
ENSMUST00000074849.6
ENSMUST00000079534.4 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr15_-_98296083 | 0.29 |
ENSMUST00000169721.1
ENSMUST00000023722.5 |
Zfp641
|
zinc finger protein 641 |
| chr19_+_4839366 | 0.28 |
ENSMUST00000088653.2
|
Ccdc87
|
coiled-coil domain containing 87 |
| chr12_+_85599047 | 0.28 |
ENSMUST00000177587.1
|
Jdp2
|
Jun dimerization protein 2 |
| chr9_+_102739648 | 0.28 |
ENSMUST00000178539.1
|
Gm5627
|
predicted gene 5627 |
| chr15_+_3270767 | 0.28 |
ENSMUST00000082424.4
ENSMUST00000159158.1 ENSMUST00000159216.1 ENSMUST00000160311.1 |
Sepp1
|
selenoprotein P, plasma, 1 |
| chr4_-_139131058 | 0.28 |
ENSMUST00000143971.1
|
Minos1
|
mitochondrial inner membrane organizing system 1 |
| chr5_+_34336289 | 0.28 |
ENSMUST00000182709.1
ENSMUST00000030992.6 |
Rnf4
|
ring finger protein 4 |
| chr1_-_14755966 | 0.28 |
ENSMUST00000027062.5
|
Msc
|
musculin |
| chr2_-_130284422 | 0.28 |
ENSMUST00000028892.4
|
Idh3b
|
isocitrate dehydrogenase 3 (NAD+) beta |
| chr2_-_30415509 | 0.28 |
ENSMUST00000134120.1
ENSMUST00000102854.3 |
Crat
|
carnitine acetyltransferase |
| chr8_+_70863127 | 0.28 |
ENSMUST00000050921.2
|
A230052G05Rik
|
RIKEN cDNA A230052G05 gene |
| chr15_-_99457742 | 0.27 |
ENSMUST00000023747.7
|
Nckap5l
|
NCK-associated protein 5-like |
| chr4_-_130574150 | 0.27 |
ENSMUST00000105993.3
|
Nkain1
|
Na+/K+ transporting ATPase interacting 1 |
| chr8_+_3665747 | 0.27 |
ENSMUST00000014118.2
|
1810033B17Rik
|
RIKEN cDNA 1810033B17 gene |
| chr11_+_69088490 | 0.27 |
ENSMUST00000021273.6
ENSMUST00000117780.1 |
Vamp2
|
vesicle-associated membrane protein 2 |
| chr2_-_10080055 | 0.27 |
ENSMUST00000130067.1
ENSMUST00000139810.1 |
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr11_-_94677404 | 0.26 |
ENSMUST00000116349.2
|
Xylt2
|
xylosyltransferase II |
| chr5_-_5694559 | 0.26 |
ENSMUST00000115426.2
|
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr9_+_44043384 | 0.26 |
ENSMUST00000114840.1
|
Thy1
|
thymus cell antigen 1, theta |
| chr16_-_20426322 | 0.26 |
ENSMUST00000115547.2
ENSMUST00000096199.4 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr4_+_128993224 | 0.25 |
ENSMUST00000030583.6
ENSMUST00000102604.4 |
Ak2
|
adenylate kinase 2 |
| chr15_+_10223974 | 0.25 |
ENSMUST00000128450.1
ENSMUST00000148257.1 ENSMUST00000128921.1 |
Prlr
|
prolactin receptor |
| chr12_+_85599388 | 0.25 |
ENSMUST00000050687.6
|
Jdp2
|
Jun dimerization protein 2 |
| chr2_+_174760619 | 0.25 |
ENSMUST00000029030.2
|
Edn3
|
endothelin 3 |
| chr11_-_99244058 | 0.25 |
ENSMUST00000103132.3
ENSMUST00000038214.6 |
Krt222
|
keratin 222 |
| chr2_+_92375306 | 0.25 |
ENSMUST00000028650.8
|
Pex16
|
peroxisomal biogenesis factor 16 |
| chr1_+_75546449 | 0.25 |
ENSMUST00000150142.1
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr2_+_178119166 | 0.25 |
ENSMUST00000108916.1
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr5_-_112252513 | 0.25 |
ENSMUST00000112385.1
|
Cryba4
|
crystallin, beta A4 |
| chr4_-_114908892 | 0.25 |
ENSMUST00000068654.3
|
Foxd2
|
forkhead box D2 |
| chr13_+_44439722 | 0.24 |
ENSMUST00000182568.1
|
1700029N11Rik
|
RIKEN cDNA 1700029N11 gene |
| chr8_+_107119110 | 0.24 |
ENSMUST00000046116.1
|
C630050I24Rik
|
RIKEN cDNA C630050I24 gene |
| chr9_+_37401993 | 0.24 |
ENSMUST00000115046.1
ENSMUST00000102895.4 |
Robo4
|
roundabout homolog 4 (Drosophila) |
| chr5_-_31047998 | 0.24 |
ENSMUST00000114665.1
ENSMUST00000006817.4 |
Slc5a6
|
solute carrier family 5 (sodium-dependent vitamin transporter), member 6 |
| chr9_-_46235260 | 0.24 |
ENSMUST00000121916.1
ENSMUST00000034586.2 |
Apoc3
|
apolipoprotein C-III |
| chr2_+_121295437 | 0.24 |
ENSMUST00000110639.1
|
Map1a
|
microtubule-associated protein 1 A |
| chr9_+_22003035 | 0.24 |
ENSMUST00000115331.2
ENSMUST00000003493.7 |
Prkcsh
|
protein kinase C substrate 80K-H |
| chr11_-_120551126 | 0.24 |
ENSMUST00000026121.2
|
Ppp1r27
|
protein phosphatase 1, regulatory subunit 27 |
| chr2_+_129593528 | 0.24 |
ENSMUST00000049262.7
ENSMUST00000163034.1 ENSMUST00000160276.1 |
Sirpa
|
signal-regulatory protein alpha |
| chr7_+_98494222 | 0.24 |
ENSMUST00000165205.1
|
Lrrc32
|
leucine rich repeat containing 32 |
| chr11_-_116843449 | 0.24 |
ENSMUST00000047616.3
|
Jmjd6
|
jumonji domain containing 6 |
| chr4_-_19922599 | 0.24 |
ENSMUST00000029900.5
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr13_-_52929458 | 0.24 |
ENSMUST00000123599.1
|
Auh
|
AU RNA binding protein/enoyl-coenzyme A hydratase |
| chr9_-_53975246 | 0.23 |
ENSMUST00000048409.7
|
Elmod1
|
ELMO/CED-12 domain containing 1 |
| chr11_-_70646972 | 0.23 |
ENSMUST00000014750.8
|
Slc25a11
|
solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Esrrb_Esrra
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.9 | 2.8 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.3 | 1.5 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.3 | 0.8 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.2 | 1.2 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 0.2 | 1.6 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 0.9 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.2 | 0.4 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.2 | 1.0 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 1.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 0.5 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 0.2 | 0.5 | GO:0070256 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.2 | 1.5 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.2 | 1.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.2 | 1.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 0.5 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 | 0.1 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.1 | 0.9 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.1 | GO:1990379 | lipid transport across blood brain barrier(GO:1990379) |
| 0.1 | 2.0 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.6 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 1.2 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.1 | 1.9 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 0.4 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 | 0.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.4 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.5 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 0.3 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) regulation of glutamine transport(GO:2000485) |
| 0.1 | 1.1 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 1.4 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 0.3 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 0.3 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.1 | GO:1900142 | regulation of oligodendrocyte apoptotic process(GO:1900141) negative regulation of oligodendrocyte apoptotic process(GO:1900142) |
| 0.1 | 0.4 | GO:1902477 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.1 | 0.3 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 1.0 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.9 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 0.6 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.6 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.3 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.3 | GO:1901526 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 0.3 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.6 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.1 | 0.2 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.3 | GO:2000612 | regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 1.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.4 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.1 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.3 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 1.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 1.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.2 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.1 | 0.4 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 0.3 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.2 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.1 | 0.3 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.1 | 0.7 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.3 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.1 | 0.7 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 0.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.1 | 1.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.1 | 0.6 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.1 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.1 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 1.2 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.1 | 0.2 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.4 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 1.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.2 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.3 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.4 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.5 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.5 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.3 | GO:0002278 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.6 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.2 | GO:0072363 | regulation of glycolytic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072363) |
| 0.0 | 0.3 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.0 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.1 | GO:0006231 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.2 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.5 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.6 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.3 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.7 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 1.9 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.1 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.4 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.1 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.3 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.7 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.4 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.2 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.0 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 1.0 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.0 | 0.1 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.0 | 0.1 | GO:1902031 | regulation of pentose-phosphate shunt(GO:0043456) regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.4 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:1990000 | amyloid fibril formation(GO:1990000) |
| 0.0 | 0.1 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.0 | 0.8 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.1 | GO:0070347 | brown fat cell proliferation(GO:0070342) regulation of brown fat cell proliferation(GO:0070347) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.4 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.1 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:1904672 | regulation of somatic stem cell population maintenance(GO:1904672) |
| 0.0 | 1.9 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.1 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.5 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.3 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.3 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 1.4 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.2 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.0 | 0.0 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.0 | 0.1 | GO:0072530 | purine-containing compound transmembrane transport(GO:0072530) |
| 0.0 | 0.1 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.0 | GO:0099624 | atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.0 | 0.2 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.3 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.3 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.1 | GO:0071281 | cellular response to iron ion(GO:0071281) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.2 | 0.5 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.2 | 1.6 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 1.6 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.9 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.7 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.3 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.1 | 1.5 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 0.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 2.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.3 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.3 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 0.8 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.4 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.2 | GO:0005940 | septin ring(GO:0005940) |
| 0.1 | 0.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.4 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 0.7 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.2 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 0.3 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.0 | 0.3 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.2 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.3 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.5 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.7 | GO:0098798 | mitochondrial protein complex(GO:0098798) |
| 0.0 | 2.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.7 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.2 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 0.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.9 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.0 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 1.0 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 1.7 | GO:0044455 | mitochondrial membrane part(GO:0044455) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.3 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 2.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 4.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 1.4 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0080130 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.6 | 1.7 | GO:0004771 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 0.4 | 1.6 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.4 | 1.4 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.3 | 1.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 0.5 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.2 | 1.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 0.5 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.2 | 1.5 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 1.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 0.4 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 1.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.7 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.4 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.6 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 1.0 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.5 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.1 | 0.3 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.1 | 0.8 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.6 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.3 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.5 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.3 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 1.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.2 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.4 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 0.5 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 1.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.2 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.1 | 0.3 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.4 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.5 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.7 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 1.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 0.3 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.2 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 0.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 1.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) serine binding(GO:0070905) |
| 0.0 | 0.2 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0033699 | DNA 5'-adenosine monophosphate hydrolase activity(GO:0033699) |
| 0.0 | 0.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.1 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.4 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.4 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.2 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.7 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 2.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 1.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.1 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.0 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 1.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 2.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 3.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 0.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 1.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 1.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 2.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |