Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for E2f4
Z-value: 4.67
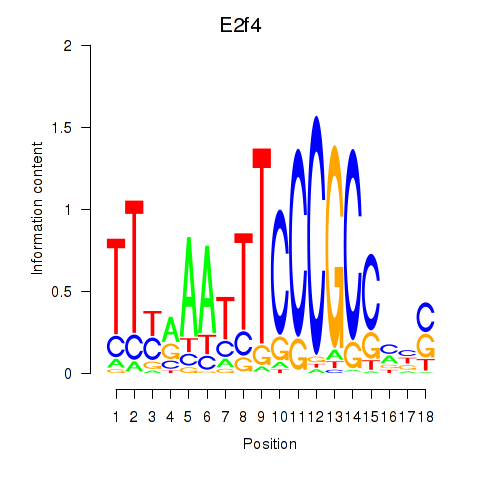
Transcription factors associated with E2f4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f4
|
ENSMUSG00000014859.8 | E2F transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f4 | mm10_v2_chr8_+_105297663_105297742 | 0.94 | 5.0e-17 | Click! |
Activity profile of E2f4 motif
Sorted Z-values of E2f4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_72438519 | 29.37 |
ENSMUST00000184604.1
|
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr9_+_72438534 | 27.71 |
ENSMUST00000034746.8
|
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr2_+_163054682 | 27.20 |
ENSMUST00000018005.3
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr4_+_134510999 | 25.35 |
ENSMUST00000105866.2
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr18_-_34751502 | 24.23 |
ENSMUST00000060710.7
|
Cdc25c
|
cell division cycle 25C |
| chr8_-_53638945 | 23.67 |
ENSMUST00000047768.4
|
Neil3
|
nei like 3 (E. coli) |
| chr14_-_47418407 | 23.55 |
ENSMUST00000043296.3
|
Dlgap5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr4_-_132345715 | 23.17 |
ENSMUST00000084250.4
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr16_-_30310773 | 22.32 |
ENSMUST00000061190.6
|
Gp5
|
glycoprotein 5 (platelet) |
| chr4_-_132345686 | 22.17 |
ENSMUST00000030726.6
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr14_-_87141114 | 22.02 |
ENSMUST00000168889.1
|
Diap3
|
diaphanous homolog 3 (Drosophila) |
| chr19_+_6084983 | 21.24 |
ENSMUST00000025704.2
|
Cdca5
|
cell division cycle associated 5 |
| chr6_+_124830217 | 20.98 |
ENSMUST00000131847.1
ENSMUST00000151674.1 |
Cdca3
|
cell division cycle associated 3 |
| chr13_+_23535411 | 20.87 |
ENSMUST00000080859.5
|
Hist1h3g
|
histone cluster 1, H3g |
| chr14_-_87141206 | 20.41 |
ENSMUST00000022599.7
|
Diap3
|
diaphanous homolog 3 (Drosophila) |
| chr10_-_88146867 | 19.31 |
ENSMUST00000164121.1
ENSMUST00000164803.1 ENSMUST00000168163.1 ENSMUST00000048518.9 |
Parpbp
|
PARP1 binding protein |
| chr1_-_169531343 | 18.41 |
ENSMUST00000028000.7
|
Nuf2
|
NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr3_-_54915867 | 17.96 |
ENSMUST00000070342.3
|
Sertm1
|
serine rich and transmembrane domain containing 1 |
| chr1_-_189688074 | 17.68 |
ENSMUST00000171929.1
ENSMUST00000165962.1 |
Cenpf
|
centromere protein F |
| chr4_-_116123618 | 17.23 |
ENSMUST00000102704.3
ENSMUST00000102705.3 |
Rad54l
|
RAD54 like (S. cerevisiae) |
| chr17_-_25727364 | 15.97 |
ENSMUST00000170070.1
ENSMUST00000048054.7 |
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 |
| chr1_-_169531447 | 15.88 |
ENSMUST00000111368.1
|
Nuf2
|
NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr11_+_98907801 | 15.22 |
ENSMUST00000092706.6
|
Cdc6
|
cell division cycle 6 |
| chr13_-_23745511 | 15.12 |
ENSMUST00000091752.2
|
Hist1h3c
|
histone cluster 1, H3c |
| chrX_+_8271381 | 14.94 |
ENSMUST00000033512.4
|
Slc38a5
|
solute carrier family 38, member 5 |
| chrX_+_8271133 | 14.82 |
ENSMUST00000127103.1
ENSMUST00000115591.1 |
Slc38a5
|
solute carrier family 38, member 5 |
| chr7_-_38107490 | 13.80 |
ENSMUST00000108023.3
|
Ccne1
|
cyclin E1 |
| chr18_-_58209926 | 13.78 |
ENSMUST00000025497.6
|
Fbn2
|
fibrillin 2 |
| chrX_+_8271642 | 13.06 |
ENSMUST00000115590.1
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr9_-_123678782 | 13.05 |
ENSMUST00000170591.1
ENSMUST00000171647.1 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr4_-_133967235 | 12.15 |
ENSMUST00000123234.1
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr4_-_133967296 | 11.50 |
ENSMUST00000105893.1
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr13_-_73937761 | 11.30 |
ENSMUST00000022053.8
|
Trip13
|
thyroid hormone receptor interactor 13 |
| chr9_-_32344237 | 11.22 |
ENSMUST00000034533.5
|
Kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr9_+_107950952 | 10.95 |
ENSMUST00000049348.3
|
Traip
|
TRAF-interacting protein |
| chr6_+_113531675 | 10.89 |
ENSMUST00000036340.5
ENSMUST00000101051.2 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr9_+_119937606 | 10.73 |
ENSMUST00000035100.5
|
Ttc21a
|
tetratricopeptide repeat domain 21A |
| chr2_+_103970221 | 10.54 |
ENSMUST00000111140.2
ENSMUST00000111139.2 |
Lmo2
|
LIM domain only 2 |
| chr15_-_76639840 | 10.37 |
ENSMUST00000166974.1
ENSMUST00000168185.1 |
Tonsl
|
tonsoku-like, DNA repair protein |
| chr11_-_101551837 | 10.23 |
ENSMUST00000017290.4
|
Brca1
|
breast cancer 1 |
| chr12_+_117843873 | 10.06 |
ENSMUST00000176735.1
ENSMUST00000177339.1 |
Cdca7l
|
cell division cycle associated 7 like |
| chr1_-_33669745 | 9.82 |
ENSMUST00000027312.9
|
Prim2
|
DNA primase, p58 subunit |
| chr1_+_86303221 | 9.68 |
ENSMUST00000113306.2
|
B3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr17_-_29237759 | 9.10 |
ENSMUST00000137727.1
ENSMUST00000024805.7 |
Cpne5
|
copine V |
| chr4_-_133967893 | 9.03 |
ENSMUST00000100472.3
ENSMUST00000136327.1 |
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr1_+_52630692 | 9.02 |
ENSMUST00000165859.1
|
Tmem194b
|
transmembrane protein 194B |
| chr7_-_92874196 | 8.62 |
ENSMUST00000032877.9
|
4632434I11Rik
|
RIKEN cDNA 4632434I11 gene |
| chr1_-_60043087 | 8.46 |
ENSMUST00000027172.6
|
Ica1l
|
islet cell autoantigen 1-like |
| chr11_+_16951371 | 8.35 |
ENSMUST00000109635.1
ENSMUST00000061327.1 |
Fbxo48
|
F-box protein 48 |
| chr18_+_34751803 | 8.26 |
ENSMUST00000181453.1
ENSMUST00000181641.1 |
2010110K18Rik
|
RIKEN cDNA 2010110K18 gene |
| chr11_-_6606053 | 8.24 |
ENSMUST00000045713.3
|
Nacad
|
NAC alpha domain containing |
| chr17_-_24251382 | 8.22 |
ENSMUST00000115390.3
|
Ccnf
|
cyclin F |
| chr18_+_56707725 | 8.14 |
ENSMUST00000025486.8
|
Lmnb1
|
lamin B1 |
| chr16_-_90727329 | 7.97 |
ENSMUST00000099554.4
|
Mis18a
|
MIS18 kinetochore protein homolog A (S. pombe) |
| chr12_+_106010263 | 7.24 |
ENSMUST00000021539.8
ENSMUST00000085026.4 ENSMUST00000072040.5 |
Vrk1
|
vaccinia related kinase 1 |
| chr8_+_18595131 | 7.20 |
ENSMUST00000039412.8
|
Mcph1
|
microcephaly, primary autosomal recessive 1 |
| chr5_+_88764983 | 7.16 |
ENSMUST00000031311.9
|
Dck
|
deoxycytidine kinase |
| chr1_-_57377476 | 6.61 |
ENSMUST00000181949.1
|
4930558J18Rik
|
RIKEN cDNA 4930558J18 gene |
| chr2_+_106695594 | 6.48 |
ENSMUST00000016530.7
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr4_-_59783800 | 6.41 |
ENSMUST00000107526.1
ENSMUST00000095063.4 |
Inip
|
INTS3 and NABP interacting protein |
| chr4_-_133967953 | 6.24 |
ENSMUST00000102553.4
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr18_-_60848911 | 6.23 |
ENSMUST00000177172.1
ENSMUST00000175934.1 ENSMUST00000176630.1 |
Tcof1
|
Treacher Collins Franceschetti syndrome 1, homolog |
| chr8_+_18595526 | 6.18 |
ENSMUST00000146819.1
|
Mcph1
|
microcephaly, primary autosomal recessive 1 |
| chr7_+_126781483 | 5.58 |
ENSMUST00000172352.1
ENSMUST00000094037.4 |
Tbx6
|
T-box 6 |
| chr10_+_79997463 | 5.57 |
ENSMUST00000171637.1
ENSMUST00000043866.7 |
Abca7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr10_-_117792663 | 5.54 |
ENSMUST00000167943.1
ENSMUST00000064848.5 |
Nup107
|
nucleoporin 107 |
| chr4_+_11558914 | 5.53 |
ENSMUST00000178703.1
ENSMUST00000095145.5 ENSMUST00000108306.2 ENSMUST00000070755.6 |
Rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr5_-_138172383 | 5.52 |
ENSMUST00000000505.9
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chrX_+_164980592 | 5.44 |
ENSMUST00000101082.4
ENSMUST00000167446.1 ENSMUST00000057150.6 |
Fancb
|
Fanconi anemia, complementation group B |
| chr4_-_44704006 | 5.26 |
ENSMUST00000146335.1
|
Pax5
|
paired box gene 5 |
| chr9_-_65580040 | 5.12 |
ENSMUST00000068944.7
|
Plekho2
|
pleckstrin homology domain containing, family O member 2 |
| chr17_+_50698525 | 5.00 |
ENSMUST00000061681.7
|
Gm7334
|
predicted gene 7334 |
| chrX_-_8074720 | 4.99 |
ENSMUST00000115636.3
ENSMUST00000115638.3 |
Suv39h1
|
suppressor of variegation 3-9 homolog 1 (Drosophila) |
| chr2_+_129593528 | 4.89 |
ENSMUST00000049262.7
ENSMUST00000163034.1 ENSMUST00000160276.1 |
Sirpa
|
signal-regulatory protein alpha |
| chr15_+_12824815 | 4.75 |
ENSMUST00000169061.1
|
Drosha
|
drosha, ribonuclease type III |
| chr15_+_12824841 | 4.72 |
ENSMUST00000090292.5
|
Drosha
|
drosha, ribonuclease type III |
| chr12_-_118198917 | 4.62 |
ENSMUST00000084806.6
|
Dnah11
|
dynein, axonemal, heavy chain 11 |
| chr6_-_56704673 | 4.59 |
ENSMUST00000170382.2
|
Lsm5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr9_+_44334685 | 4.52 |
ENSMUST00000052686.2
|
H2afx
|
H2A histone family, member X |
| chrX_+_162901226 | 4.51 |
ENSMUST00000101095.2
|
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chrX_+_105079735 | 4.42 |
ENSMUST00000033577.4
|
Pbdc1
|
polysaccharide biosynthesis domain containing 1 |
| chr2_-_166713758 | 4.34 |
ENSMUST00000036719.5
|
Prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr1_-_77515048 | 4.34 |
ENSMUST00000027451.6
|
Epha4
|
Eph receptor A4 |
| chr2_+_30286406 | 4.23 |
ENSMUST00000138666.1
ENSMUST00000113634.2 |
Nup188
|
nucleoporin 188 |
| chrX_-_167209149 | 4.16 |
ENSMUST00000112176.1
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr12_+_99884498 | 4.15 |
ENSMUST00000153627.1
|
Tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr2_+_129593195 | 4.03 |
ENSMUST00000099113.3
ENSMUST00000103202.3 |
Sirpa
|
signal-regulatory protein alpha |
| chr9_+_44134562 | 3.95 |
ENSMUST00000034650.8
ENSMUST00000098852.2 |
Mcam
|
melanoma cell adhesion molecule |
| chr5_-_138171813 | 3.93 |
ENSMUST00000155902.1
ENSMUST00000148879.1 |
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr5_-_25705791 | 3.81 |
ENSMUST00000030773.7
|
Xrcc2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr7_+_58658181 | 3.74 |
ENSMUST00000168747.1
|
Atp10a
|
ATPase, class V, type 10A |
| chr14_-_54517353 | 3.67 |
ENSMUST00000023873.5
|
Prmt5
|
protein arginine N-methyltransferase 5 |
| chrX_+_162901567 | 3.65 |
ENSMUST00000112303.1
ENSMUST00000033727.7 |
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chr9_-_83441065 | 3.64 |
ENSMUST00000034791.8
ENSMUST00000034793.8 |
Lca5
|
Leber congenital amaurosis 5 (human) |
| chr5_-_33652296 | 3.61 |
ENSMUST00000151081.1
ENSMUST00000101354.3 |
Slbp
|
stem-loop binding protein |
| chr10_+_127677064 | 3.47 |
ENSMUST00000118612.1
ENSMUST00000048099.4 |
Tmem194
|
transmembrane protein 194 |
| chr2_+_181319714 | 3.43 |
ENSMUST00000098971.4
ENSMUST00000054622.8 ENSMUST00000108814.1 ENSMUST00000048608.9 ENSMUST00000108815.1 |
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr7_-_127208423 | 3.41 |
ENSMUST00000120705.1
|
Tbc1d10b
|
TBC1 domain family, member 10b |
| chr17_+_87975044 | 3.36 |
ENSMUST00000005503.3
|
Msh6
|
mutS homolog 6 (E. coli) |
| chr5_-_33652339 | 3.28 |
ENSMUST00000075670.6
|
Slbp
|
stem-loop binding protein |
| chr2_+_181319806 | 3.23 |
ENSMUST00000153112.1
|
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr1_+_9798123 | 3.16 |
ENSMUST00000168907.1
ENSMUST00000166384.1 |
Sgk3
|
serum/glucocorticoid regulated kinase 3 |
| chrX_-_78583882 | 3.14 |
ENSMUST00000114025.1
ENSMUST00000134602.1 ENSMUST00000114024.2 |
Prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr19_+_46075842 | 3.13 |
ENSMUST00000165017.1
|
Nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr7_+_97371604 | 3.12 |
ENSMUST00000098300.4
|
Alg8
|
asparagine-linked glycosylation 8 (alpha-1,3-glucosyltransferase) |
| chr10_+_88146992 | 3.10 |
ENSMUST00000052355.7
|
Nup37
|
nucleoporin 37 |
| chr2_+_153649617 | 3.03 |
ENSMUST00000109771.1
|
Dnmt3b
|
DNA methyltransferase 3B |
| chr4_-_58553311 | 2.96 |
ENSMUST00000107571.1
ENSMUST00000055018.4 |
Lpar1
|
lysophosphatidic acid receptor 1 |
| chrX_+_162901762 | 2.87 |
ENSMUST00000112302.1
ENSMUST00000112301.1 |
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chr4_-_58553553 | 2.86 |
ENSMUST00000107575.2
ENSMUST00000107574.1 ENSMUST00000147354.1 |
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr9_+_57708534 | 2.80 |
ENSMUST00000043990.7
ENSMUST00000142807.1 |
Edc3
|
enhancer of mRNA decapping 3 homolog (S. cerevisiae) |
| chr14_+_76488436 | 2.77 |
ENSMUST00000101618.2
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr5_+_30889326 | 2.77 |
ENSMUST00000124908.1
|
Agbl5
|
ATP/GTP binding protein-like 5 |
| chr11_+_108425192 | 2.70 |
ENSMUST00000150863.2
ENSMUST00000061287.5 ENSMUST00000149683.2 |
Cep112
|
centrosomal protein 112 |
| chr4_-_43562397 | 2.68 |
ENSMUST00000030187.7
|
Tln1
|
talin 1 |
| chr10_+_88147061 | 2.65 |
ENSMUST00000169309.1
|
Nup37
|
nucleoporin 37 |
| chr2_+_129592914 | 2.65 |
ENSMUST00000103203.1
|
Sirpa
|
signal-regulatory protein alpha |
| chr19_-_5366626 | 2.49 |
ENSMUST00000025762.8
|
Banf1
|
barrier to autointegration factor 1 |
| chr5_+_141241490 | 2.45 |
ENSMUST00000085774.4
|
Sdk1
|
sidekick homolog 1 (chicken) |
| chr17_-_35046726 | 2.31 |
ENSMUST00000097338.4
|
Msh5
|
mutS homolog 5 (E. coli) |
| chr13_+_73937799 | 2.27 |
ENSMUST00000099384.2
|
Brd9
|
bromodomain containing 9 |
| chr12_-_4233958 | 2.15 |
ENSMUST00000111169.3
ENSMUST00000020981.5 |
Cenpo
|
centromere protein O |
| chr1_+_134415414 | 2.04 |
ENSMUST00000112237.1
|
Adipor1
|
adiponectin receptor 1 |
| chr15_-_98831498 | 2.03 |
ENSMUST00000168846.1
|
Prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr9_-_82975475 | 2.00 |
ENSMUST00000034787.5
|
Phip
|
pleckstrin homology domain interacting protein |
| chr7_+_6383310 | 1.87 |
ENSMUST00000081022.7
|
Zfp28
|
zinc finger protein 28 |
| chr17_+_29490812 | 1.87 |
ENSMUST00000024811.6
|
Pim1
|
proviral integration site 1 |
| chrX_-_78583782 | 1.76 |
ENSMUST00000177904.1
|
Prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr8_+_124023394 | 1.67 |
ENSMUST00000034457.8
|
Urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr17_-_17624458 | 1.66 |
ENSMUST00000041047.2
|
Lnpep
|
leucyl/cystinyl aminopeptidase |
| chr9_-_60688118 | 1.52 |
ENSMUST00000114034.2
ENSMUST00000065603.5 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr4_-_3835595 | 1.49 |
ENSMUST00000138502.1
|
Rps20
|
ribosomal protein S20 |
| chr2_-_3512746 | 1.45 |
ENSMUST00000056700.7
ENSMUST00000027961.5 |
Hspa14
Hspa14
|
heat shock protein 14 heat shock protein 14 |
| chr4_+_84884418 | 1.44 |
ENSMUST00000169371.2
|
Cntln
|
centlein, centrosomal protein |
| chr5_+_21372642 | 1.35 |
ENSMUST00000035799.5
|
Fgl2
|
fibrinogen-like protein 2 |
| chr7_+_24884651 | 1.33 |
ENSMUST00000153451.2
ENSMUST00000108429.1 |
Rps19
|
ribosomal protein S19 |
| chr6_+_70726430 | 1.25 |
ENSMUST00000103410.1
|
Igkc
|
immunoglobulin kappa constant |
| chr14_+_55578123 | 1.24 |
ENSMUST00000174484.1
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chrX_-_37110257 | 1.15 |
ENSMUST00000076265.6
|
Upf3b
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr17_+_34982099 | 1.13 |
ENSMUST00000007266.7
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr11_-_106999369 | 1.10 |
ENSMUST00000106768.1
ENSMUST00000144834.1 |
Kpna2
|
karyopherin (importin) alpha 2 |
| chr4_+_44004438 | 1.09 |
ENSMUST00000107846.3
|
Clta
|
clathrin, light polypeptide (Lca) |
| chr4_-_132463873 | 1.08 |
ENSMUST00000102567.3
|
Med18
|
mediator of RNA polymerase II transcription, subunit 18 homolog (yeast) |
| chr1_+_133351909 | 1.05 |
ENSMUST00000112287.1
|
Ren1
|
renin 1 structural |
| chr4_+_84884276 | 1.04 |
ENSMUST00000047023.6
|
Cntln
|
centlein, centrosomal protein |
| chr11_-_106999482 | 1.02 |
ENSMUST00000018506.6
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chr5_-_92348871 | 1.00 |
ENSMUST00000038816.6
ENSMUST00000118006.1 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr7_+_24884611 | 0.89 |
ENSMUST00000108428.1
|
Rps19
|
ribosomal protein S19 |
| chr4_-_58553184 | 0.84 |
ENSMUST00000145361.1
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr7_-_16285454 | 0.83 |
ENSMUST00000174270.1
|
Ccdc9
|
coiled-coil domain containing 9 |
| chr1_+_33669816 | 0.82 |
ENSMUST00000051203.5
|
1700001G17Rik
|
RIKEN cDNA 1700001G17 gene |
| chr7_-_16286010 | 0.80 |
ENSMUST00000145519.2
|
Ccdc9
|
coiled-coil domain containing 9 |
| chr17_+_34982154 | 0.79 |
ENSMUST00000173004.1
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr2_-_145935014 | 0.77 |
ENSMUST00000001818.4
|
Crnkl1
|
Crn, crooked neck-like 1 (Drosophila) |
| chr7_-_109865586 | 0.77 |
ENSMUST00000007423.5
ENSMUST00000106728.2 ENSMUST00000106729.1 |
Scube2
|
signal peptide, CUB domain, EGF-like 2 |
| chrY_+_90784738 | 0.73 |
ENSMUST00000179483.1
|
Erdr1
|
erythroid differentiation regulator 1 |
| chr1_+_179803376 | 0.69 |
ENSMUST00000097454.2
|
Gm10518
|
predicted gene 10518 |
| chr3_-_142881942 | 0.68 |
ENSMUST00000043812.8
|
Pkn2
|
protein kinase N2 |
| chr17_-_87025353 | 0.66 |
ENSMUST00000024957.6
|
Pigf
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr11_+_26593241 | 0.63 |
ENSMUST00000089614.4
|
Gm6899
|
predicted gene 6899 |
| chrX_+_170009892 | 0.49 |
ENSMUST00000180251.1
|
Gm21887
|
predicted gene, 21887 |
| chrX_+_71981522 | 0.48 |
ENSMUST00000114569.1
|
Fate1
|
fetal and adult testis expressed 1 |
| chr5_+_30888852 | 0.47 |
ENSMUST00000069705.4
ENSMUST00000031057.8 ENSMUST00000046182.5 ENSMUST00000114704.1 ENSMUST00000061213.6 ENSMUST00000114696.1 ENSMUST00000114700.2 |
Agbl5
|
ATP/GTP binding protein-like 5 |
| chr13_-_69533839 | 0.45 |
ENSMUST00000044081.7
|
Papd7
|
PAP associated domain containing 7 |
| chr2_+_174076296 | 0.45 |
ENSMUST00000155000.1
ENSMUST00000134876.1 ENSMUST00000147038.1 |
Stx16
|
syntaxin 16 |
| chr1_+_157412352 | 0.38 |
ENSMUST00000061537.5
|
2810025M15Rik
|
RIKEN cDNA 2810025M15 gene |
| chr17_-_84187939 | 0.36 |
ENSMUST00000060366.6
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr7_+_97579868 | 0.25 |
ENSMUST00000042399.7
ENSMUST00000107153.1 |
Rsf1
|
remodeling and spacing factor 1 |
| chr2_+_71786923 | 0.23 |
ENSMUST00000112101.1
ENSMUST00000028522.3 |
Itga6
|
integrin alpha 6 |
| chr9_+_92250039 | 0.22 |
ENSMUST00000093801.3
|
Plscr1
|
phospholipid scramblase 1 |
| chr9_+_118926453 | 0.19 |
ENSMUST00000073109.5
|
Ctdspl
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr17_-_35046539 | 0.15 |
ENSMUST00000007250.7
|
Msh5
|
mutS homolog 5 (E. coli) |
| chr2_+_145934800 | 0.09 |
ENSMUST00000138774.1
ENSMUST00000152515.1 ENSMUST00000130168.1 ENSMUST00000133433.1 ENSMUST00000118002.1 |
4930529M08Rik
|
RIKEN cDNA 4930529M08 gene |
| chrX_+_105079761 | 0.01 |
ENSMUST00000119477.1
|
Pbdc1
|
polysaccharide biosynthesis domain containing 1 |
| chr9_+_99456243 | 0.01 |
ENSMUST00000163199.2
|
1600029I14Rik
|
RIKEN cDNA 1600029I14 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 21.2 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 3.4 | 10.2 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 3.3 | 13.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 2.9 | 54.7 | GO:0070986 | left/right axis specification(GO:0070986) |
| 2.9 | 42.8 | GO:0015816 | glycine transport(GO:0015816) |
| 2.8 | 13.8 | GO:0071692 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 2.7 | 8.1 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 2.5 | 9.8 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 2.2 | 13.4 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 1.9 | 5.6 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 1.8 | 5.5 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 1.8 | 7.2 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 1.8 | 7.2 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 1.8 | 16.0 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 1.8 | 24.7 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 1.7 | 6.7 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 1.7 | 6.7 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 1.6 | 11.2 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) potassium ion import across plasma membrane(GO:1990573) |
| 1.4 | 4.3 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 1.4 | 11.3 | GO:0007144 | female meiosis I(GO:0007144) |
| 1.4 | 9.5 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 1.3 | 38.9 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 1.3 | 8.0 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 1.3 | 3.8 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 1.2 | 8.2 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 1.1 | 66.2 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 1.1 | 10.9 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 1.0 | 38.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 1.0 | 3.0 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.9 | 32.5 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.9 | 26.1 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.9 | 22.3 | GO:0010544 | negative regulation of platelet activation(GO:0010544) |
| 0.8 | 4.2 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.8 | 24.2 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.8 | 5.6 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.8 | 11.0 | GO:0006241 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.7 | 3.7 | GO:0060161 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) histone H4-R3 methylation(GO:0043985) positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.7 | 11.0 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.7 | 10.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.7 | 6.9 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.6 | 10.7 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.6 | 5.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.6 | 17.7 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.5 | 27.2 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.5 | 6.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.5 | 4.6 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.5 | 23.7 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.5 | 10.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.5 | 5.0 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.4 | 4.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.4 | 5.3 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.4 | 9.1 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.3 | 2.8 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.3 | 4.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.3 | 2.7 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.3 | 12.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.3 | 2.5 | GO:0015074 | DNA integration(GO:0015074) |
| 0.3 | 3.2 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.3 | 1.9 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.3 | 1.0 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.2 | 23.2 | GO:0007051 | spindle organization(GO:0007051) |
| 0.2 | 2.0 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.2 | 1.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.2 | 2.5 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 3.1 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.2 | 0.8 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.2 | 4.5 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.2 | 2.1 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.2 | 15.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.2 | 1.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 1.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 2.0 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 1.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 3.6 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 2.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 2.5 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.1 | 3.2 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 4.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.2 | GO:2000371 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.1 | 0.4 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.1 | 6.4 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.1 | 2.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 3.7 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 3.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 5.7 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 1.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 3.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 1.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 3.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 34.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 5.4 | 27.2 | GO:0031523 | Myb complex(GO:0031523) |
| 3.5 | 10.4 | GO:0035101 | FACT complex(GO:0035101) |
| 3.2 | 9.5 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 2.4 | 21.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 2.0 | 9.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 1.8 | 16.0 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 1.7 | 10.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 1.5 | 17.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 1.3 | 6.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 1.2 | 10.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 1.2 | 8.1 | GO:0005638 | lamin filament(GO:0005638) |
| 1.1 | 23.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 1.1 | 11.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 1.0 | 5.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 1.0 | 6.9 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.9 | 15.8 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.9 | 6.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.8 | 4.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.8 | 13.8 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.8 | 3.8 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.7 | 9.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.6 | 20.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.5 | 15.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.5 | 57.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.5 | 5.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.5 | 3.4 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.4 | 3.7 | GO:0034709 | methylosome(GO:0034709) |
| 0.4 | 47.8 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.4 | 15.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.4 | 1.1 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
| 0.3 | 1.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.3 | 25.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.3 | 13.4 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.2 | 13.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 5.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 14.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 15.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 8.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 10.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.8 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.5 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 7.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 2.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 3.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 4.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 2.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 3.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 11.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 5.3 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 20.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 6.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 3.6 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 1.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 12.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 2.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 16.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 6.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 9.8 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.1 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 14.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 45.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 4.7 | 23.7 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 3.7 | 11.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 3.1 | 42.8 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 2.8 | 11.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 2.5 | 27.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 2.3 | 6.9 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 2.1 | 6.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 1.9 | 9.5 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 1.9 | 13.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 1.9 | 5.6 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 1.8 | 7.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 1.7 | 17.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 1.5 | 16.0 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 1.2 | 7.2 | GO:0035184 | nucleosomal histone binding(GO:0031493) histone threonine kinase activity(GO:0035184) |
| 1.1 | 3.4 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) single guanine insertion binding(GO:0032142) |
| 1.0 | 3.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 1.0 | 6.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.9 | 3.7 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.8 | 5.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.8 | 36.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.7 | 4.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.6 | 3.8 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.6 | 17.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.6 | 5.0 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.5 | 2.0 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.5 | 24.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.5 | 17.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.5 | 2.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.4 | 3.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.4 | 9.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.4 | 13.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.4 | 9.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.3 | 2.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 8.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.3 | 9.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.3 | 14.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.3 | 13.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 20.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 16.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 9.7 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.2 | 1.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 0.8 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.2 | 1.9 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.2 | 4.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 3.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 3.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 4.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 11.6 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.1 | 2.8 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.1 | 3.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 4.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 2.0 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 1.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 1.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 14.2 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.1 | 0.5 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 1.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 10.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 2.2 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 1.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 2.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 8.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 8.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.7 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 2.5 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 10.1 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 25.0 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 2.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 59.8 | PID ATR PATHWAY | ATR signaling pathway |
| 1.0 | 13.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.8 | 33.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.5 | 16.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.4 | 47.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.4 | 30.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.3 | 10.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.3 | 10.4 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.3 | 11.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 8.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 4.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 6.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 2.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 5.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 9.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 15.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 4.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 24.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 2.2 | 56.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 1.5 | 22.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 1.4 | 9.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 1.2 | 10.9 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.1 | 15.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 1.1 | 9.8 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.9 | 42.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.8 | 40.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.6 | 9.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.6 | 11.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.6 | 6.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.6 | 1.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.5 | 60.9 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.5 | 9.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.4 | 10.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.4 | 11.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.4 | 7.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 9.7 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.3 | 2.5 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.3 | 8.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 5.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 2.7 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.2 | 17.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 11.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 3.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.2 | 4.2 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 2.5 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.1 | 2.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 3.1 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 3.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 4.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 7.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 4.3 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 1.1 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.2 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |