Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Dlx1
Z-value: 3.53
Transcription factors associated with Dlx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Dlx1
|
ENSMUSG00000041911.3 | distal-less homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dlx1 | mm10_v2_chr2_+_71528657_71528683 | 0.42 | 1.1e-02 | Click! |
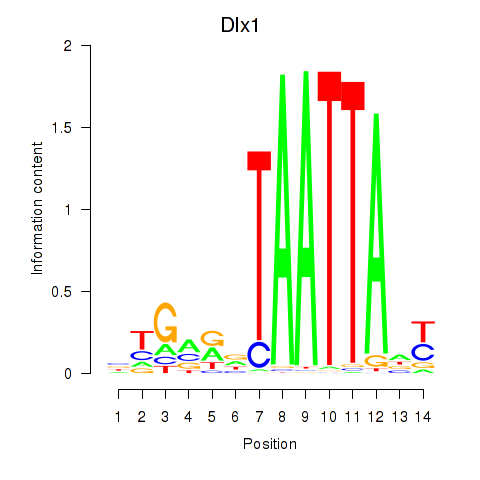
Activity profile of Dlx1 motif
Sorted Z-values of Dlx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_84988194 | 25.05 |
ENSMUST00000028466.5
|
Prg3
|
proteoglycan 3 |
| chr19_-_34166037 | 23.70 |
ENSMUST00000025686.7
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr11_+_87793470 | 22.49 |
ENSMUST00000020779.4
|
Mpo
|
myeloperoxidase |
| chr14_+_80000292 | 19.08 |
ENSMUST00000088735.3
|
Olfm4
|
olfactomedin 4 |
| chr1_+_131638306 | 17.67 |
ENSMUST00000073350.6
|
Ctse
|
cathepsin E |
| chr11_+_58918004 | 17.64 |
ENSMUST00000108818.3
ENSMUST00000020792.5 |
Btnl10
|
butyrophilin-like 10 |
| chr16_+_36277145 | 15.53 |
ENSMUST00000042097.9
|
Stfa1
|
stefin A1 |
| chr1_+_40439767 | 15.06 |
ENSMUST00000173514.1
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr17_+_40811089 | 14.71 |
ENSMUST00000024721.7
|
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr11_+_58917889 | 14.58 |
ENSMUST00000069941.6
|
Btnl10
|
butyrophilin-like 10 |
| chr14_+_27000362 | 13.90 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr4_+_34893772 | 13.23 |
ENSMUST00000029975.3
ENSMUST00000135871.1 ENSMUST00000108130.1 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr1_+_40439627 | 12.03 |
ENSMUST00000097772.3
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr1_-_132390301 | 11.91 |
ENSMUST00000132435.1
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr2_+_131491764 | 10.77 |
ENSMUST00000028806.5
ENSMUST00000110179.2 ENSMUST00000110189.2 ENSMUST00000110182.2 ENSMUST00000110183.2 ENSMUST00000110186.2 ENSMUST00000110188.1 |
Smox
|
spermine oxidase |
| chr6_+_41354105 | 10.72 |
ENSMUST00000072103.5
|
Try10
|
trypsin 10 |
| chr6_+_30541582 | 10.57 |
ENSMUST00000096066.4
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr9_-_123678782 | 10.44 |
ENSMUST00000170591.1
ENSMUST00000171647.1 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr5_-_108749448 | 10.26 |
ENSMUST00000068946.7
|
Rnf212
|
ring finger protein 212 |
| chr6_-_40999479 | 10.25 |
ENSMUST00000166306.1
|
Gm2663
|
predicted gene 2663 |
| chr1_+_107535508 | 10.12 |
ENSMUST00000182198.1
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr2_-_126500631 | 9.93 |
ENSMUST00000129187.1
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr3_-_75270073 | 9.79 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr17_-_48432723 | 9.73 |
ENSMUST00000046549.3
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr8_-_85380964 | 9.27 |
ENSMUST00000122452.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr5_+_90768511 | 9.25 |
ENSMUST00000031319.6
|
Ppbp
|
pro-platelet basic protein |
| chr12_+_109540979 | 9.08 |
ENSMUST00000129245.1
ENSMUST00000143836.1 ENSMUST00000124106.1 |
Meg3
|
maternally expressed 3 |
| chr17_+_5799491 | 9.07 |
ENSMUST00000181484.1
|
3300005D01Rik
|
RIKEN cDNA 3300005D01 gene |
| chr7_-_119459266 | 9.04 |
ENSMUST00000033255.5
|
Gp2
|
glycoprotein 2 (zymogen granule membrane) |
| chr13_+_104229366 | 8.89 |
ENSMUST00000022227.6
|
Cenpk
|
centromere protein K |
| chr9_-_123678873 | 8.82 |
ENSMUST00000040960.6
|
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr7_-_133702515 | 8.76 |
ENSMUST00000153698.1
|
Uros
|
uroporphyrinogen III synthase |
| chr14_+_32321987 | 8.69 |
ENSMUST00000022480.7
|
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr8_-_4779513 | 8.47 |
ENSMUST00000022945.7
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr19_-_40588374 | 8.01 |
ENSMUST00000175932.1
ENSMUST00000176955.1 ENSMUST00000149476.2 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr19_+_58759700 | 7.82 |
ENSMUST00000026081.3
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr5_-_43981757 | 7.81 |
ENSMUST00000061299.7
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr1_-_93342734 | 7.81 |
ENSMUST00000027493.3
|
Pask
|
PAS domain containing serine/threonine kinase |
| chr8_+_94172618 | 7.69 |
ENSMUST00000034214.6
|
Mt2
|
metallothionein 2 |
| chr3_-_113258837 | 7.64 |
ENSMUST00000098673.3
|
Amy2a5
|
amylase 2a5 |
| chr16_-_75909272 | 7.63 |
ENSMUST00000114239.2
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr3_-_20275659 | 7.59 |
ENSMUST00000011607.5
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr15_+_44457522 | 7.48 |
ENSMUST00000166957.1
ENSMUST00000038336.5 |
Pkhd1l1
|
polycystic kidney and hepatic disease 1-like 1 |
| chr19_-_40588338 | 7.47 |
ENSMUST00000176939.1
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr2_+_119047116 | 7.42 |
ENSMUST00000152380.1
ENSMUST00000099542.2 |
Casc5
|
cancer susceptibility candidate 5 |
| chr15_-_36879816 | 7.18 |
ENSMUST00000100713.2
|
Gm10384
|
predicted gene 10384 |
| chr15_+_84232030 | 7.12 |
ENSMUST00000023072.6
|
Parvb
|
parvin, beta |
| chr11_+_83437678 | 6.89 |
ENSMUST00000037378.4
|
1700020L24Rik
|
RIKEN cDNA 1700020L24 gene |
| chr6_-_36811361 | 6.87 |
ENSMUST00000101534.1
|
Ptn
|
pleiotrophin |
| chr3_-_116253467 | 6.86 |
ENSMUST00000090473.5
|
Gpr88
|
G-protein coupled receptor 88 |
| chr10_+_115817247 | 6.73 |
ENSMUST00000035563.7
ENSMUST00000080630.3 ENSMUST00000179196.1 |
Tspan8
|
tetraspanin 8 |
| chr9_+_65890237 | 6.68 |
ENSMUST00000045802.6
|
2810417H13Rik
|
RIKEN cDNA 2810417H13 gene |
| chrX_+_150547375 | 6.65 |
ENSMUST00000066337.6
ENSMUST00000112715.1 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr17_-_35027909 | 6.58 |
ENSMUST00000040151.2
|
Sapcd1
|
suppressor APC domain containing 1 |
| chr8_+_23411490 | 6.51 |
ENSMUST00000033952.7
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr3_-_14778452 | 6.42 |
ENSMUST00000094365.4
|
Car1
|
carbonic anhydrase 1 |
| chr11_+_69045640 | 6.40 |
ENSMUST00000108666.1
ENSMUST00000021277.5 |
Aurkb
|
aurora kinase B |
| chr14_+_118787894 | 6.22 |
ENSMUST00000047761.6
ENSMUST00000071546.7 |
Cldn10
|
claudin 10 |
| chr9_-_58741543 | 6.17 |
ENSMUST00000098674.4
|
2410076I21Rik
|
RIKEN cDNA 2410076I21 gene |
| chr18_-_74207771 | 6.06 |
ENSMUST00000040188.8
ENSMUST00000177604.1 |
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr1_+_139454747 | 6.01 |
ENSMUST00000053364.8
ENSMUST00000097554.3 |
Aspm
|
asp (abnormal spindle)-like, microcephaly associated (Drosophila) |
| chr9_-_95750335 | 5.98 |
ENSMUST00000053785.3
|
Trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr2_-_121235689 | 5.96 |
ENSMUST00000142400.1
|
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr6_+_123172893 | 5.91 |
ENSMUST00000088455.5
|
Clec4b2
|
C-type lectin domain family 4, member b2 |
| chr12_-_101028983 | 5.86 |
ENSMUST00000068411.3
ENSMUST00000085096.3 |
Ccdc88c
|
coiled-coil domain containing 88C |
| chr11_-_106314494 | 5.84 |
ENSMUST00000167143.1
|
Cd79b
|
CD79B antigen |
| chr3_-_113291449 | 5.79 |
ENSMUST00000179568.1
|
Amy2a4
|
amylase 2a4 |
| chr9_-_21963568 | 5.65 |
ENSMUST00000006397.5
|
Epor
|
erythropoietin receptor |
| chr7_+_43427622 | 5.64 |
ENSMUST00000177164.2
|
Lim2
|
lens intrinsic membrane protein 2 |
| chr13_-_23551648 | 5.52 |
ENSMUST00000102971.1
|
Hist1h4f
|
histone cluster 1, H4f |
| chr16_+_32756336 | 5.49 |
ENSMUST00000135753.1
|
Muc4
|
mucin 4 |
| chr11_+_11487671 | 5.48 |
ENSMUST00000020410.4
|
4930415F15Rik
|
RIKEN cDNA 4930415F15 gene |
| chr12_-_115790884 | 5.35 |
ENSMUST00000081809.5
|
Ighv1-73
|
immunoglobulin heavy variable 1-73 |
| chr8_+_116921735 | 5.33 |
ENSMUST00000034205.4
|
Cenpn
|
centromere protein N |
| chr9_+_119063429 | 5.27 |
ENSMUST00000141185.1
ENSMUST00000126251.1 ENSMUST00000136561.1 |
Vill
|
villin-like |
| chr14_+_118854695 | 5.25 |
ENSMUST00000100314.3
|
Cldn10
|
claudin 10 |
| chr8_-_62123106 | 5.15 |
ENSMUST00000034052.6
ENSMUST00000034054.7 |
Anxa10
|
annexin A10 |
| chr6_-_129275360 | 5.14 |
ENSMUST00000032259.3
|
Cd69
|
CD69 antigen |
| chr1_-_126830632 | 5.13 |
ENSMUST00000112583.1
ENSMUST00000094609.3 |
Nckap5
|
NCK-associated protein 5 |
| chr10_+_119992916 | 5.08 |
ENSMUST00000105261.2
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr2_+_109280738 | 5.03 |
ENSMUST00000028527.7
|
Kif18a
|
kinesin family member 18A |
| chr13_+_52596847 | 4.98 |
ENSMUST00000055087.6
|
Syk
|
spleen tyrosine kinase |
| chr12_-_55014329 | 4.97 |
ENSMUST00000172875.1
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr12_+_108605757 | 4.96 |
ENSMUST00000109854.2
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chrX_-_74246534 | 4.96 |
ENSMUST00000101454.2
ENSMUST00000033699.6 |
Flna
|
filamin, alpha |
| chr10_-_6980376 | 4.90 |
ENSMUST00000105617.1
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr10_-_62527438 | 4.88 |
ENSMUST00000160987.1
|
Srgn
|
serglycin |
| chr10_-_62527413 | 4.81 |
ENSMUST00000160643.1
|
Srgn
|
serglycin |
| chr13_+_44729535 | 4.80 |
ENSMUST00000174068.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr3_-_106219477 | 4.74 |
ENSMUST00000082219.5
|
Chi3l4
|
chitinase 3-like 4 |
| chr11_+_74082907 | 4.66 |
ENSMUST00000178159.1
|
Zfp616
|
zinc finger protein 616 |
| chr7_-_25005895 | 4.64 |
ENSMUST00000102858.3
ENSMUST00000080882.6 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr5_-_30945393 | 4.59 |
ENSMUST00000031051.6
|
Cgref1
|
cell growth regulator with EF hand domain 1 |
| chr10_+_119992962 | 4.55 |
ENSMUST00000154238.1
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr13_+_23544052 | 4.44 |
ENSMUST00000075558.2
|
Hist1h3f
|
histone cluster 1, H3f |
| chrX_-_160906998 | 4.38 |
ENSMUST00000069417.5
|
Gja6
|
gap junction protein, alpha 6 |
| chr6_-_122340525 | 4.35 |
ENSMUST00000112600.2
|
Phc1
|
polyhomeotic-like 1 (Drosophila) |
| chrX_-_53114530 | 4.29 |
ENSMUST00000114843.2
|
Plac1
|
placental specific protein 1 |
| chr2_+_85979312 | 4.28 |
ENSMUST00000170610.1
|
Olfr1030
|
olfactory receptor 1030 |
| chr3_-_10254213 | 4.27 |
ENSMUST00000172126.1
ENSMUST00000117917.1 |
Fabp12
|
fatty acid binding protein 12 |
| chr10_-_62422427 | 4.25 |
ENSMUST00000020277.8
|
Hkdc1
|
hexokinase domain containing 1 |
| chr4_-_132075250 | 4.19 |
ENSMUST00000105970.1
ENSMUST00000105975.1 |
Epb4.1
|
erythrocyte protein band 4.1 |
| chr5_+_141856692 | 4.15 |
ENSMUST00000074546.6
|
Sdk1
|
sidekick homolog 1 (chicken) |
| chr2_+_84839395 | 4.15 |
ENSMUST00000146816.1
ENSMUST00000028469.7 |
Slc43a1
|
solute carrier family 43, member 1 |
| chr1_-_172895048 | 4.07 |
ENSMUST00000027824.5
|
Apcs
|
serum amyloid P-component |
| chr17_+_34914459 | 4.06 |
ENSMUST00000007249.8
|
Slc44a4
|
solute carrier family 44, member 4 |
| chr6_-_122340499 | 4.05 |
ENSMUST00000160843.1
|
Phc1
|
polyhomeotic-like 1 (Drosophila) |
| chr13_-_95525239 | 4.03 |
ENSMUST00000022185.8
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr2_-_160619971 | 4.03 |
ENSMUST00000109473.1
|
Gm14221
|
predicted gene 14221 |
| chr15_-_34356421 | 4.01 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr8_+_119700009 | 3.98 |
ENSMUST00000095171.3
|
Atp2c2
|
ATPase, Ca++ transporting, type 2C, member 2 |
| chr2_+_119047129 | 3.98 |
ENSMUST00000153300.1
ENSMUST00000028799.5 |
Casc5
|
cancer susceptibility candidate 5 |
| chr14_+_61309753 | 3.94 |
ENSMUST00000055159.7
|
Arl11
|
ADP-ribosylation factor-like 11 |
| chr3_-_27153861 | 3.94 |
ENSMUST00000108300.1
ENSMUST00000108298.2 |
Ect2
|
ect2 oncogene |
| chr6_-_50456085 | 3.87 |
ENSMUST00000146341.1
ENSMUST00000071728.4 |
Osbpl3
|
oxysterol binding protein-like 3 |
| chr7_+_141746736 | 3.84 |
ENSMUST00000026590.8
|
Muc2
|
mucin 2 |
| chr19_-_32196393 | 3.72 |
ENSMUST00000151822.1
|
Sgms1
|
sphingomyelin synthase 1 |
| chr12_-_87444017 | 3.71 |
ENSMUST00000091090.4
|
2700073G19Rik
|
RIKEN cDNA 2700073G19 gene |
| chr9_-_57836706 | 3.71 |
ENSMUST00000164010.1
ENSMUST00000171444.1 ENSMUST00000098686.3 |
Arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr10_-_76110956 | 3.69 |
ENSMUST00000120757.1
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr17_-_26099257 | 3.69 |
ENSMUST00000053575.3
|
Gm8186
|
predicted gene 8186 |
| chr14_-_104522615 | 3.66 |
ENSMUST00000022716.2
|
Rnf219
|
ring finger protein 219 |
| chr19_+_55180799 | 3.65 |
ENSMUST00000025936.5
|
Tectb
|
tectorin beta |
| chr7_+_45621805 | 3.61 |
ENSMUST00000033100.4
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr11_-_69895523 | 3.57 |
ENSMUST00000001631.6
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr2_+_4300462 | 3.57 |
ENSMUST00000175669.1
|
Frmd4a
|
FERM domain containing 4A |
| chr16_-_69863718 | 3.52 |
ENSMUST00000076500.7
|
Speer2
|
spermatogenesis associated glutamate (E)-rich protein 2 |
| chr6_+_72097561 | 3.51 |
ENSMUST00000069994.4
ENSMUST00000114112.1 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr13_-_58354862 | 3.41 |
ENSMUST00000043605.5
|
Kif27
|
kinesin family member 27 |
| chr19_-_55241236 | 3.40 |
ENSMUST00000069183.6
|
Gucy2g
|
guanylate cyclase 2g |
| chr3_+_146121655 | 3.35 |
ENSMUST00000039450.4
|
Mcoln3
|
mucolipin 3 |
| chr6_+_122513643 | 3.31 |
ENSMUST00000118626.1
|
Mfap5
|
microfibrillar associated protein 5 |
| chr2_-_73453918 | 3.30 |
ENSMUST00000102679.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chrX_-_57338598 | 3.28 |
ENSMUST00000033468.4
ENSMUST00000114736.1 |
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr19_+_34100943 | 3.27 |
ENSMUST00000025685.6
|
Lipm
|
lipase, family member M |
| chr9_-_36726374 | 3.25 |
ENSMUST00000172702.2
ENSMUST00000172742.1 ENSMUST00000034625.5 |
Chek1
|
checkpoint kinase 1 |
| chr6_-_129876659 | 3.24 |
ENSMUST00000014687.4
ENSMUST00000122219.1 |
Klra17
|
killer cell lectin-like receptor, subfamily A, member 17 |
| chr17_+_8236031 | 3.24 |
ENSMUST00000164411.2
|
Ccr6
|
chemokine (C-C motif) receptor 6 |
| chr18_-_23981555 | 3.19 |
ENSMUST00000115829.1
|
Zscan30
|
zinc finger and SCAN domain containing 30 |
| chr3_-_17230976 | 3.19 |
ENSMUST00000177874.1
|
Gm5283
|
predicted gene 5283 |
| chr19_-_50678642 | 3.16 |
ENSMUST00000072685.6
ENSMUST00000164039.2 |
Sorcs1
|
VPS10 domain receptor protein SORCS 1 |
| chr16_+_92478743 | 3.16 |
ENSMUST00000160494.1
|
2410124H12Rik
|
RIKEN cDNA 2410124H12 gene |
| chr9_-_21312255 | 3.15 |
ENSMUST00000115433.3
ENSMUST00000003397.7 |
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr4_+_145585166 | 3.15 |
ENSMUST00000105739.1
ENSMUST00000119718.1 |
Gm13212
|
predicted gene 13212 |
| chr10_-_129902726 | 3.13 |
ENSMUST00000071557.1
|
Olfr815
|
olfactory receptor 815 |
| chr6_+_122513583 | 3.08 |
ENSMUST00000032210.7
ENSMUST00000148517.1 |
Mfap5
|
microfibrillar associated protein 5 |
| chr19_-_11604828 | 3.02 |
ENSMUST00000025582.4
|
Ms4a6d
|
membrane-spanning 4-domains, subfamily A, member 6D |
| chr19_-_38043559 | 3.02 |
ENSMUST00000041475.8
ENSMUST00000172095.2 |
Myof
|
myoferlin |
| chr7_+_27452417 | 3.01 |
ENSMUST00000108357.1
|
Blvrb
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr4_-_41464816 | 3.00 |
ENSMUST00000108055.2
ENSMUST00000154535.1 ENSMUST00000030148.5 |
Kif24
|
kinesin family member 24 |
| chr2_-_85675173 | 2.99 |
ENSMUST00000099917.1
|
Olfr1006
|
olfactory receptor 1006 |
| chr18_+_44104407 | 2.97 |
ENSMUST00000081271.5
|
Spink12
|
serine peptidase inhibitor, Kazal type 11 |
| chr16_-_3908596 | 2.97 |
ENSMUST00000123235.2
|
1700037C18Rik
|
RIKEN cDNA 1700037C18 gene |
| chr18_+_37518341 | 2.96 |
ENSMUST00000097609.1
|
Pcdhb22
|
protocadherin beta 22 |
| chr19_+_53140430 | 2.96 |
ENSMUST00000111741.2
|
Add3
|
adducin 3 (gamma) |
| chr13_+_23555023 | 2.95 |
ENSMUST00000045301.6
|
Hist1h1d
|
histone cluster 1, H1d |
| chr12_+_102128718 | 2.95 |
ENSMUST00000159329.1
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr5_+_100845709 | 2.95 |
ENSMUST00000144623.1
|
Agpat9
|
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr1_+_86021935 | 2.95 |
ENSMUST00000052854.6
ENSMUST00000125083.1 ENSMUST00000152501.1 ENSMUST00000113344.1 ENSMUST00000130504.1 ENSMUST00000153247.2 |
Spata3
|
spermatogenesis associated 3 |
| chr2_-_113848655 | 2.93 |
ENSMUST00000102545.1
ENSMUST00000110948.1 |
Arhgap11a
|
Rho GTPase activating protein 11A |
| chr1_-_172027251 | 2.93 |
ENSMUST00000138714.1
|
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chr12_+_55124528 | 2.91 |
ENSMUST00000177768.1
|
Fam177a
|
family with sequence similarity 177, member A |
| chr2_-_164171113 | 2.91 |
ENSMUST00000045196.3
|
Kcns1
|
K+ voltage-gated channel, subfamily S, 1 |
| chr9_-_90255927 | 2.89 |
ENSMUST00000144646.1
|
Tbc1d2b
|
TBC1 domain family, member 2B |
| chr3_-_130730375 | 2.86 |
ENSMUST00000079085.6
|
Rpl34
|
ribosomal protein L34 |
| chr15_+_92344359 | 2.76 |
ENSMUST00000181901.1
|
Gm26760
|
predicted gene, 26760 |
| chr10_+_80264942 | 2.76 |
ENSMUST00000105362.1
ENSMUST00000105361.3 |
Dazap1
|
DAZ associated protein 1 |
| chr2_-_34913976 | 2.76 |
ENSMUST00000028232.3
|
Phf19
|
PHD finger protein 19 |
| chr2_+_91650116 | 2.70 |
ENSMUST00000111331.2
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr1_+_58210397 | 2.69 |
ENSMUST00000040442.5
|
Aox4
|
aldehyde oxidase 4 |
| chrX_+_153139941 | 2.65 |
ENSMUST00000039720.4
ENSMUST00000144175.2 |
Rragb
|
Ras-related GTP binding B |
| chr4_+_12906838 | 2.65 |
ENSMUST00000143186.1
ENSMUST00000183345.1 |
Triqk
|
triple QxxK/R motif containing |
| chr13_+_23575753 | 2.64 |
ENSMUST00000105105.1
|
Hist1h3d
|
histone cluster 1, H3d |
| chr7_-_101869307 | 2.64 |
ENSMUST00000140584.1
ENSMUST00000134145.1 |
Folr1
|
folate receptor 1 (adult) |
| chr6_-_128275577 | 2.62 |
ENSMUST00000130454.1
|
Tead4
|
TEA domain family member 4 |
| chr6_+_78370877 | 2.60 |
ENSMUST00000096904.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr4_-_154928187 | 2.59 |
ENSMUST00000123514.1
|
Tnfrsf14
|
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
| chr4_+_101507947 | 2.56 |
ENSMUST00000149047.1
ENSMUST00000106929.3 |
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr17_-_47016956 | 2.54 |
ENSMUST00000165525.1
|
Gm16494
|
predicted gene 16494 |
| chrY_+_90785442 | 2.51 |
ENSMUST00000177591.1
ENSMUST00000177671.1 ENSMUST00000179077.1 |
Erdr1
|
erythroid differentiation regulator 1 |
| chr5_+_19907502 | 2.51 |
ENSMUST00000101558.3
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr13_-_62371936 | 2.50 |
ENSMUST00000107989.3
|
Gm3604
|
predicted gene 3604 |
| chr10_-_88605017 | 2.46 |
ENSMUST00000119185.1
ENSMUST00000121629.1 |
Mybpc1
|
myosin binding protein C, slow-type |
| chrX_-_164250368 | 2.45 |
ENSMUST00000112263.1
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr2_-_120154600 | 2.45 |
ENSMUST00000028755.7
|
Ehd4
|
EH-domain containing 4 |
| chrX_-_8074720 | 2.44 |
ENSMUST00000115636.3
ENSMUST00000115638.3 |
Suv39h1
|
suppressor of variegation 3-9 homolog 1 (Drosophila) |
| chr3_+_59952185 | 2.43 |
ENSMUST00000094227.3
|
Gm9696
|
predicted gene 9696 |
| chr16_+_38346986 | 2.38 |
ENSMUST00000050273.8
ENSMUST00000120495.1 ENSMUST00000119704.1 |
Cox17
Gm21987
|
cytochrome c oxidase assembly protein 17 predicted gene 21987 |
| chr17_-_31129602 | 2.38 |
ENSMUST00000024827.4
|
Tff3
|
trefoil factor 3, intestinal |
| chr10_-_51631458 | 2.36 |
ENSMUST00000020062.3
|
Gprc6a
|
G protein-coupled receptor, family C, group 6, member A |
| chr4_+_146654927 | 2.36 |
ENSMUST00000070932.3
|
Gm13248
|
predicted gene 13248 |
| chr12_-_12940600 | 2.36 |
ENSMUST00000130990.1
|
Mycn
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) |
| chr2_+_69219971 | 2.35 |
ENSMUST00000005364.5
ENSMUST00000112317.2 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr8_-_21456560 | 2.34 |
ENSMUST00000110752.3
|
Gm7849
|
predicted gene 7849 |
| chr9_-_62026788 | 2.34 |
ENSMUST00000034817.4
|
Paqr5
|
progestin and adipoQ receptor family member V |
| chr4_+_62525369 | 2.33 |
ENSMUST00000062145.1
|
4933430I17Rik
|
RIKEN cDNA 4933430I17 gene |
| chr12_+_84069325 | 2.31 |
ENSMUST00000046422.4
ENSMUST00000072505.4 |
Acot5
|
acyl-CoA thioesterase 5 |
| chr19_+_23723279 | 2.30 |
ENSMUST00000067077.1
|
Gm9938
|
predicted gene 9938 |
| chr7_+_84853573 | 2.29 |
ENSMUST00000078172.4
|
Olfr291
|
olfactory receptor 291 |
| chr17_+_17316078 | 2.28 |
ENSMUST00000105311.3
|
Gm6712
|
predicted gene 6712 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Dlx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 22.5 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 6.5 | 26.1 | GO:0045575 | basophil activation(GO:0045575) |
| 5.2 | 15.5 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 4.9 | 14.7 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 4.8 | 19.3 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 2.6 | 28.8 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 2.4 | 9.7 | GO:0033380 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 2.4 | 2.4 | GO:1904732 | regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 2.4 | 7.1 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 2.3 | 6.9 | GO:0061743 | motor learning(GO:0061743) |
| 2.2 | 8.8 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 2.2 | 6.5 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 2.1 | 6.4 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 2.1 | 10.3 | GO:0006311 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 1.7 | 6.9 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 1.7 | 5.1 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 1.7 | 5.0 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 1.5 | 9.3 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 1.5 | 10.8 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 1.5 | 14.9 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 1.4 | 5.5 | GO:0002838 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) |
| 1.4 | 1.4 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 1.4 | 4.1 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 1.3 | 4.0 | GO:1900135 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 1.2 | 5.0 | GO:0032672 | regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) cellular response to molecule of fungal origin(GO:0071226) |
| 1.2 | 3.6 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 1.2 | 6.0 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 1.2 | 20.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 1.1 | 7.7 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 1.0 | 1.0 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 1.0 | 7.8 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 1.0 | 9.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.9 | 13.2 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.9 | 1.8 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.8 | 4.2 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.8 | 5.0 | GO:0032439 | endosome localization(GO:0032439) negative regulation of vacuolar transport(GO:1903336) |
| 0.8 | 3.2 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.7 | 4.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.7 | 2.2 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.7 | 2.2 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.7 | 4.3 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.7 | 7.8 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.7 | 4.2 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.7 | 2.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.6 | 1.3 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.6 | 2.3 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.6 | 7.4 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.5 | 2.7 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.5 | 2.6 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.5 | 3.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.5 | 6.0 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.5 | 1.9 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.5 | 4.6 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.5 | 9.3 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.4 | 7.6 | GO:0019377 | glycolipid catabolic process(GO:0019377) |
| 0.4 | 4.9 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.4 | 4.8 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.4 | 2.6 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.4 | 2.6 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.4 | 2.6 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.4 | 1.3 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.4 | 3.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.4 | 5.6 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.4 | 1.6 | GO:0015827 | angiotensin-mediated drinking behavior(GO:0003051) aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.4 | 6.0 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.4 | 2.8 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.4 | 1.2 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.4 | 5.0 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.4 | 3.0 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.4 | 5.9 | GO:0002579 | positive regulation of antigen processing and presentation(GO:0002579) |
| 0.4 | 3.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.4 | 1.8 | GO:0015819 | lysine transport(GO:0015819) |
| 0.4 | 3.2 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.4 | 1.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.4 | 1.1 | GO:0061738 | mitotic cytokinesis checkpoint(GO:0044878) late endosomal microautophagy(GO:0061738) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.3 | 1.0 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.3 | 5.5 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.3 | 4.8 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 | 2.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.3 | 8.9 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.3 | 8.5 | GO:0097435 | fibril organization(GO:0097435) |
| 0.3 | 1.2 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.3 | 16.9 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.3 | 3.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.3 | 3.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.3 | 2.6 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.3 | 1.7 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.3 | 2.9 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.3 | 0.6 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.3 | 1.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.3 | 0.8 | GO:1904742 | protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.3 | 2.4 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.3 | 1.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.3 | 3.7 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.3 | 1.0 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.3 | 1.0 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.3 | 0.8 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.2 | 7.6 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.2 | 0.5 | GO:1900239 | regulation of phenotypic switching(GO:1900239) |
| 0.2 | 2.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.2 | 6.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 3.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 1.4 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.2 | 3.9 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.2 | 0.7 | GO:0061193 | taste bud development(GO:0061193) |
| 0.2 | 1.8 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.2 | 1.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.2 | 1.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.2 | 2.4 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 0.4 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.2 | 0.4 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) CD4-positive, alpha-beta T cell proliferation(GO:0035739) |
| 0.2 | 5.3 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.2 | 0.8 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.2 | 11.9 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.2 | 1.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.2 | 4.3 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.2 | 11.2 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.2 | 2.4 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.2 | 5.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.2 | 4.1 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.2 | 0.7 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.2 | 0.7 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.2 | 1.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 2.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 0.6 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 8.6 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.2 | 1.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.7 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 1.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.4 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.1 | 1.3 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.6 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 | 2.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 3.3 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 1.3 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 1.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) vesicle uncoating(GO:0072319) |
| 0.1 | 8.4 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 2.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.1 | 6.3 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 4.1 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 1.0 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.1 | 3.4 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 | 5.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 2.9 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 4.7 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 3.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 2.4 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 0.6 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 10.5 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 1.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 1.2 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 | 2.3 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 2.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 1.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 3.4 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.1 | 1.0 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 10.8 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.1 | 0.2 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 3.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 10.1 | GO:0031109 | microtubule polymerization or depolymerization(GO:0031109) |
| 0.1 | 3.5 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.1 | 1.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 2.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 7.3 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.1 | 3.4 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.1 | 5.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 1.3 | GO:0002504 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 | 1.1 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 0.3 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.1 | 0.4 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.1 | 1.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 2.2 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.1 | 0.5 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.1 | 0.9 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 2.5 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 12.4 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 1.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 2.4 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 2.2 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 4.2 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.7 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.3 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.6 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 2.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 4.6 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 2.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.4 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 1.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 3.2 | GO:0060998 | regulation of dendritic spine development(GO:0060998) |
| 0.0 | 0.9 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.6 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 1.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 1.3 | GO:0030804 | positive regulation of cyclic nucleotide biosynthetic process(GO:0030804) |
| 0.0 | 1.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 1.3 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 2.0 | GO:0034101 | erythrocyte homeostasis(GO:0034101) |
| 0.0 | 0.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 2.2 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.8 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.3 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.0 | 1.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.0 | GO:1904923 | regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) |
| 0.0 | 1.6 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.1 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 19.1 | GO:0042581 | specific granule(GO:0042581) |
| 1.5 | 10.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 1.4 | 22.5 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 1.3 | 5.1 | GO:0060187 | cell pole(GO:0060187) |
| 1.2 | 5.0 | GO:0008623 | CHRAC(GO:0008623) |
| 1.2 | 6.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 1.1 | 6.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.0 | 5.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.9 | 9.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.9 | 1.7 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.8 | 8.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.8 | 4.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.8 | 5.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.8 | 3.9 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.7 | 5.0 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.7 | 7.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.6 | 8.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.6 | 7.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.5 | 2.7 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.5 | 4.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.5 | 8.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.5 | 2.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.5 | 1.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.4 | 1.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.4 | 1.8 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.4 | 3.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.4 | 1.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.3 | 3.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.3 | 5.3 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.3 | 2.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.3 | 7.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 2.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 9.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 1.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 0.9 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 4.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 1.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 5.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 7.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 0.8 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 4.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 2.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 0.6 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 1.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 6.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 2.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 1.7 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 2.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.7 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 3.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.9 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 1.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.9 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 20.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 5.9 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 2.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 3.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.6 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 41.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 6.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 11.2 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 11.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 10.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.6 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 1.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 3.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.8 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 25.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 0.5 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 5.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 2.0 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 5.3 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 10.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 2.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 2.0 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 7.0 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.4 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 2.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 4.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 8.5 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 2.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.0 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.9 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 3.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 2.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.5 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.0 | 6.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 4.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 25.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 8.2 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 3.9 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.0 | 27.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 5.2 | 15.5 | GO:0019202 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 3.6 | 10.8 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 2.3 | 6.9 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 2.2 | 8.6 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 1.9 | 13.4 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 1.8 | 20.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 1.7 | 5.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 1.6 | 9.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 1.5 | 8.8 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 1.4 | 5.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 1.3 | 9.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.2 | 8.7 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 1.1 | 3.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 1.1 | 8.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.0 | 4.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.0 | 6.0 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 1.0 | 5.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.8 | 4.2 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.8 | 2.4 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.8 | 14.7 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.8 | 3.0 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.7 | 3.7 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.7 | 2.2 | GO:0000401 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.7 | 9.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.6 | 17.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.6 | 20.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.6 | 1.8 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.6 | 5.0 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.5 | 2.7 | GO:0016726 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.5 | 12.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.5 | 2.5 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.5 | 7.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.5 | 2.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.5 | 4.3 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.5 | 6.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.5 | 3.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.4 | 1.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.4 | 6.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 4.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.4 | 2.8 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.4 | 3.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.4 | 24.3 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.4 | 5.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 1.8 | GO:0015189 | L-ornithine transmembrane transporter activity(GO:0000064) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.4 | 6.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.3 | 1.0 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.3 | 2.0 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.3 | 3.0 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.3 | 1.6 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.3 | 9.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.3 | 3.7 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.3 | 2.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.3 | 2.4 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.3 | 1.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.3 | 9.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.3 | 5.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.3 | 1.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.3 | 2.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.3 | 1.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.2 | 2.0 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 1.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.2 | 6.7 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.2 | 0.7 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.2 | 5.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 2.6 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.2 | 1.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 7.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 2.0 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 1.5 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.2 | 4.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 1.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 1.0 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.2 | 2.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.2 | 0.7 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.2 | 1.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 1.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 1.0 | GO:0032405 | MutLalpha complex binding(GO:0032405) MutSalpha complex binding(GO:0032407) |
| 0.2 | 8.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 8.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 0.7 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.2 | 2.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 1.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 0.6 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 1.9 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 6.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 7.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.0 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 3.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.8 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 1.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 3.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.6 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 3.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 1.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 9.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.7 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 2.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 4.8 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 5.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 4.1 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 4.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.6 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 11.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.7 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 1.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 1.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 3.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 7.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 1.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 2.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 8.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 0.5 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 15.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 9.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 1.1 | GO:0043495 | adenylate cyclase binding(GO:0008179) protein anchor(GO:0043495) |
| 0.1 | 0.6 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 4.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.8 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.4 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 3.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 4.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 2.0 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 9.6 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.1 | 9.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 2.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 12.0 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 2.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 12.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 4.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 3.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 16.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 3.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.2 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.8 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 4.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 1.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 4.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.0 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.9 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.0 | 0.4 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 4.6 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 34.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.5 | 4.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 22.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.4 | 6.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 5.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.2 | 6.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 6.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 6.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.2 | 4.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 8.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 7.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 2.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 3.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 5.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 4.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 5.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 1.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 3.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 9.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 4.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 29.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 9.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 20.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 3.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 3.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 3.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 3.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 1.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 2.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 2.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 1.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 13.4 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 2.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 3.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 13.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.8 | 18.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.6 | 10.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.6 | 15.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.6 | 19.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.6 | 31.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.5 | 3.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.5 | 10.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.4 | 4.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 10.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.4 | 8.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.4 | 18.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.3 | 5.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 11.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 10.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 4.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 3.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 19.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 5.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 20.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 6.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 9.0 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.1 | 1.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 4.8 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 1.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 2.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 1.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 10.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 4.8 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 1.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.9 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 6.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 2.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 2.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 2.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.0 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATION OF FGFR SIGNALING | Genes involved in Negative regulation of FGFR signaling |
| 0.0 | 0.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 2.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |