Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Creb1
Z-value: 2.19
Transcription factors associated with Creb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Creb1
|
ENSMUSG00000025958.8 | cAMP responsive element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb1 | mm10_v2_chr1_+_64532790_64532815 | -0.11 | 5.4e-01 | Click! |
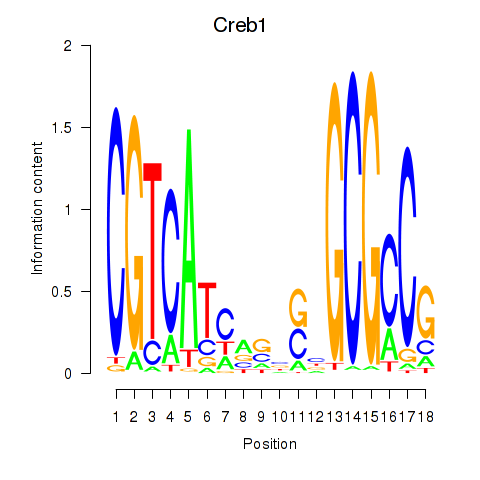
Activity profile of Creb1 motif
Sorted Z-values of Creb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_55099417 | 5.08 |
ENSMUST00000061856.5
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr1_+_185363076 | 3.19 |
ENSMUST00000046514.7
|
Eprs
|
glutamyl-prolyl-tRNA synthetase |
| chr12_-_4907705 | 2.84 |
ENSMUST00000020962.5
|
Ubxn2a
|
UBX domain protein 2A |
| chr10_+_62920630 | 2.78 |
ENSMUST00000044977.3
|
Slc25a16
|
solute carrier family 25 (mitochondrial carrier, Graves disease autoantigen), member 16 |
| chr12_+_108334341 | 2.61 |
ENSMUST00000021684.4
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr13_-_29984219 | 2.58 |
ENSMUST00000146092.1
|
E2f3
|
E2F transcription factor 3 |
| chr14_-_59597836 | 2.46 |
ENSMUST00000167100.1
ENSMUST00000022555.4 ENSMUST00000056997.7 ENSMUST00000171683.1 |
Cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr10_+_62920648 | 2.36 |
ENSMUST00000144459.1
|
Slc25a16
|
solute carrier family 25 (mitochondrial carrier, Graves disease autoantigen), member 16 |
| chr12_-_4907229 | 2.34 |
ENSMUST00000142867.1
|
Ubxn2a
|
UBX domain protein 2A |
| chr11_-_86807624 | 2.27 |
ENSMUST00000018569.7
|
Dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr8_+_40511769 | 2.24 |
ENSMUST00000098817.2
|
Vps37a
|
vacuolar protein sorting 37A (yeast) |
| chr8_+_3621529 | 2.03 |
ENSMUST00000156380.2
|
Pet100
|
PET100 homolog (S. cerevisiae) |
| chr11_-_83302586 | 2.01 |
ENSMUST00000176374.1
|
Pex12
|
peroxisomal biogenesis factor 12 |
| chr13_+_58402546 | 1.97 |
ENSMUST00000042450.8
|
Rmi1
|
RMI1, RecQ mediated genome instability 1, homolog (S. cerevisiae) |
| chrX_-_56598069 | 1.95 |
ENSMUST00000059899.2
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr1_+_180851131 | 1.91 |
ENSMUST00000038091.6
|
Sde2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr12_+_33429605 | 1.88 |
ENSMUST00000020877.7
|
Twistnb
|
TWIST neighbor |
| chr1_+_78657825 | 1.87 |
ENSMUST00000035779.8
|
Acsl3
|
acyl-CoA synthetase long-chain family member 3 |
| chr12_+_116281180 | 1.87 |
ENSMUST00000100986.2
|
Esyt2
|
extended synaptotagmin-like protein 2 |
| chr12_+_28751798 | 1.81 |
ENSMUST00000035657.7
|
Tssc1
|
tumor suppressing subtransferable candidate 1 |
| chr10_+_7792891 | 1.75 |
ENSMUST00000015901.4
|
Ppil4
|
peptidylprolyl isomerase (cyclophilin)-like 4 |
| chr12_-_57546121 | 1.67 |
ENSMUST00000044380.6
|
Foxa1
|
forkhead box A1 |
| chr7_+_46796088 | 1.66 |
ENSMUST00000006774.4
ENSMUST00000165031.1 |
Gtf2h1
|
general transcription factor II H, polypeptide 1 |
| chr1_+_43933980 | 1.62 |
ENSMUST00000087933.3
|
Tpp2
|
tripeptidyl peptidase II |
| chr18_+_9958147 | 1.58 |
ENSMUST00000025137.7
|
Thoc1
|
THO complex 1 |
| chr5_-_112343010 | 1.56 |
ENSMUST00000146510.1
ENSMUST00000031289.6 |
Srrd
|
SRR1 domain containing |
| chr16_+_22857845 | 1.55 |
ENSMUST00000004574.7
ENSMUST00000178320.1 ENSMUST00000166487.2 |
Dnajb11
|
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr10_+_44268328 | 1.51 |
ENSMUST00000039286.4
|
Atg5
|
autophagy related 5 |
| chr8_-_22653406 | 1.51 |
ENSMUST00000033938.5
|
Polb
|
polymerase (DNA directed), beta |
| chr8_-_79711631 | 1.50 |
ENSMUST00000080536.6
|
Abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr2_-_119156824 | 1.49 |
ENSMUST00000135419.1
ENSMUST00000129351.1 ENSMUST00000139519.1 ENSMUST00000094695.5 |
Rmdn3
|
regulator of microtubule dynamics 3 |
| chr8_-_40511298 | 1.45 |
ENSMUST00000149992.1
|
Cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr1_+_78657874 | 1.42 |
ENSMUST00000134566.1
ENSMUST00000142704.1 ENSMUST00000053760.5 |
Acsl3
Utp14b
|
acyl-CoA synthetase long-chain family member 3 UTP14, U3 small nucleolar ribonucleoprotein, homolog B (yeast) |
| chr12_+_51377560 | 1.37 |
ENSMUST00000021335.5
|
Scfd1
|
Sec1 family domain containing 1 |
| chr13_+_98263242 | 1.37 |
ENSMUST00000022164.8
ENSMUST00000150352.1 |
Ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr14_-_31251194 | 1.36 |
ENSMUST00000022459.3
|
Phf7
|
PHD finger protein 7 |
| chr2_+_144556229 | 1.33 |
ENSMUST00000143573.1
ENSMUST00000028916.8 ENSMUST00000155258.1 |
Sec23b
|
SEC23B (S. cerevisiae) |
| chr7_-_118533298 | 1.32 |
ENSMUST00000098090.3
ENSMUST00000032887.3 |
Coq7
|
demethyl-Q 7 |
| chr4_-_149307506 | 1.31 |
ENSMUST00000055647.8
ENSMUST00000030806.5 ENSMUST00000060537.6 |
Kif1b
|
kinesin family member 1B |
| chr12_-_56535047 | 1.30 |
ENSMUST00000178477.2
|
Nkx2-1
|
NK2 homeobox 1 |
| chr8_-_40511663 | 1.30 |
ENSMUST00000135269.1
ENSMUST00000034012.3 |
Cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr12_-_108179237 | 1.30 |
ENSMUST00000071095.7
|
Setd3
|
SET domain containing 3 |
| chr12_-_101083653 | 1.30 |
ENSMUST00000048305.8
ENSMUST00000163095.1 |
Smek1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr9_-_96478660 | 1.29 |
ENSMUST00000057500.4
|
Rnf7
|
ring finger protein 7 |
| chr8_+_60993189 | 1.28 |
ENSMUST00000034065.7
ENSMUST00000120689.1 |
Nek1
|
NIMA (never in mitosis gene a)-related expressed kinase 1 |
| chr11_+_95384662 | 1.27 |
ENSMUST00000021243.7
ENSMUST00000146556.1 |
Slc35b1
|
solute carrier family 35, member B1 |
| chr6_-_4086914 | 1.26 |
ENSMUST00000049166.4
|
Bet1
|
blocked early in transport 1 homolog (S. cerevisiae) |
| chr13_+_96542727 | 1.23 |
ENSMUST00000077672.4
ENSMUST00000109444.2 |
Col4a3bp
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
| chrX_+_71816758 | 1.23 |
ENSMUST00000114576.2
ENSMUST00000114575.3 |
Vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr9_-_96478596 | 1.22 |
ENSMUST00000071301.4
|
Rnf7
|
ring finger protein 7 |
| chr9_-_53248106 | 1.20 |
ENSMUST00000065630.6
|
Ddx10
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
| chr18_+_6765171 | 1.20 |
ENSMUST00000097680.5
|
Rab18
|
RAB18, member RAS oncogene family |
| chr1_-_134361933 | 1.18 |
ENSMUST00000049470.4
|
Tmem183a
|
transmembrane protein 183A |
| chr1_+_58973521 | 1.18 |
ENSMUST00000114296.1
ENSMUST00000027185.4 |
Stradb
|
STE20-related kinase adaptor beta |
| chr4_+_131843459 | 1.17 |
ENSMUST00000030742.4
ENSMUST00000137321.1 |
Mecr
|
mitochondrial trans-2-enoyl-CoA reductase |
| chr13_+_98263187 | 1.17 |
ENSMUST00000091356.3
ENSMUST00000123924.1 |
Ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr15_-_96460838 | 1.16 |
ENSMUST00000047835.6
|
Scaf11
|
SR-related CTD-associated factor 11 |
| chr3_+_138143483 | 1.16 |
ENSMUST00000162864.1
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr5_-_65335597 | 1.14 |
ENSMUST00000172660.1
ENSMUST00000172732.1 ENSMUST00000031092.8 |
Rfc1
|
replication factor C (activator 1) 1 |
| chr1_-_37430071 | 1.13 |
ENSMUST00000027286.6
|
Coa5
|
cytochrome C oxidase assembly factor 5 |
| chr1_-_150392719 | 1.13 |
ENSMUST00000006167.6
ENSMUST00000094477.2 ENSMUST00000097547.3 |
BC003331
|
cDNA sequence BC003331 |
| chr4_-_40279389 | 1.13 |
ENSMUST00000108108.2
ENSMUST00000095128.3 |
Ndufb6
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6 |
| chr15_+_36174010 | 1.12 |
ENSMUST00000180159.1
ENSMUST00000057177.6 |
Polr2k
|
polymerase (RNA) II (DNA directed) polypeptide K |
| chr3_-_107696462 | 1.10 |
ENSMUST00000029490.8
|
Ahcyl1
|
S-adenosylhomocysteine hydrolase-like 1 |
| chr12_+_8208107 | 1.09 |
ENSMUST00000037383.5
ENSMUST00000169104.1 |
1110057K04Rik
|
RIKEN cDNA 1110057K04 gene |
| chr5_+_67260794 | 1.09 |
ENSMUST00000161369.1
|
Tmem33
|
transmembrane protein 33 |
| chrX_-_74023745 | 1.08 |
ENSMUST00000114353.3
ENSMUST00000101458.2 |
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr19_+_8735808 | 1.08 |
ENSMUST00000049424.9
|
Wdr74
|
WD repeat domain 74 |
| chr10_+_94688739 | 1.07 |
ENSMUST00000020212.4
|
Ccdc41
|
coiled-coil domain containing 41 |
| chr13_-_96542479 | 1.07 |
ENSMUST00000022172.4
|
Polk
|
polymerase (DNA directed), kappa |
| chr14_+_54883377 | 1.07 |
ENSMUST00000022806.3
ENSMUST00000172844.1 ENSMUST00000133397.2 ENSMUST00000134077.1 |
Bcl2l2
Gm20521
|
BCL2-like 2 predicted gene 20521 |
| chr17_+_46496753 | 1.06 |
ENSMUST00000046497.6
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr12_-_73286698 | 1.06 |
ENSMUST00000116420.2
|
Trmt5
|
TRM5 tRNA methyltransferase 5 |
| chr16_+_43889936 | 1.05 |
ENSMUST00000151183.1
|
2610015P09Rik
|
RIKEN cDNA 2610015P09 gene |
| chr3_+_138143429 | 1.05 |
ENSMUST00000040321.6
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr1_-_106796687 | 1.04 |
ENSMUST00000094646.5
|
Vps4b
|
vacuolar protein sorting 4b (yeast) |
| chr5_+_124439891 | 1.03 |
ENSMUST00000059580.4
|
Setd8
|
SET domain containing (lysine methyltransferase) 8 |
| chr13_+_98263105 | 1.03 |
ENSMUST00000150916.1
|
Ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr19_-_24961545 | 1.03 |
ENSMUST00000025815.8
|
Cbwd1
|
COBW domain containing 1 |
| chr15_-_81399594 | 1.02 |
ENSMUST00000023039.8
|
St13
|
suppression of tumorigenicity 13 |
| chr4_+_115737738 | 1.01 |
ENSMUST00000106525.2
|
Efcab14
|
EF-hand calcium binding domain 14 |
| chr4_-_147904680 | 1.01 |
ENSMUST00000105716.2
ENSMUST00000105715.1 ENSMUST00000105714.1 ENSMUST00000030884.3 |
Mfn2
|
mitofusin 2 |
| chr4_-_19708922 | 1.01 |
ENSMUST00000108246.2
|
Wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr4_-_14826582 | 1.00 |
ENSMUST00000117268.1
|
Otud6b
|
OTU domain containing 6B |
| chr9_-_122950922 | 1.00 |
ENSMUST00000118422.1
|
1110059G10Rik
|
RIKEN cDNA 1110059G10 gene |
| chr4_+_115737754 | 1.00 |
ENSMUST00000106522.2
|
Efcab14
|
EF-hand calcium binding domain 14 |
| chr15_-_94589870 | 0.99 |
ENSMUST00000023087.6
ENSMUST00000152590.1 |
Twf1
|
twinfilin, actin-binding protein, homolog 1 (Drosophila) |
| chr3_-_127553233 | 0.98 |
ENSMUST00000029588.5
|
Larp7
|
La ribonucleoprotein domain family, member 7 |
| chr16_-_97922582 | 0.98 |
ENSMUST00000170757.1
|
C2cd2
|
C2 calcium-dependent domain containing 2 |
| chr1_-_150393024 | 0.97 |
ENSMUST00000097546.2
ENSMUST00000111913.2 |
BC003331
|
cDNA sequence BC003331 |
| chr11_+_59202480 | 0.95 |
ENSMUST00000108786.1
ENSMUST00000108787.2 ENSMUST00000045697.5 ENSMUST00000108785.1 ENSMUST00000108784.3 |
Mrpl55
|
mitochondrial ribosomal protein L55 |
| chr9_-_122950960 | 0.94 |
ENSMUST00000035154.3
|
1110059G10Rik
|
RIKEN cDNA 1110059G10 gene |
| chr13_-_58402533 | 0.94 |
ENSMUST00000176207.1
|
Hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr7_-_110844350 | 0.92 |
ENSMUST00000177462.1
ENSMUST00000176746.1 ENSMUST00000177236.1 |
Rnf141
|
ring finger protein 141 |
| chr12_-_56345862 | 0.92 |
ENSMUST00000021416.7
|
Mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr9_+_121710389 | 0.91 |
ENSMUST00000035113.9
|
Deb1
|
differentially expressed in B16F10 1 |
| chr13_+_96542602 | 0.90 |
ENSMUST00000179226.1
|
Col4a3bp
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
| chr10_+_82954344 | 0.90 |
ENSMUST00000095396.3
|
Gm4799
|
predicted gene 4799 |
| chr11_-_115276973 | 0.90 |
ENSMUST00000021078.2
|
Fdxr
|
ferredoxin reductase |
| chr14_+_55745678 | 0.90 |
ENSMUST00000019441.8
|
Nop9
|
NOP9 nucleolar protein |
| chr5_+_24100578 | 0.90 |
ENSMUST00000030841.5
ENSMUST00000163409.1 |
Klhl7
|
kelch-like 7 |
| chr5_+_38039224 | 0.90 |
ENSMUST00000031008.6
ENSMUST00000042146.8 ENSMUST00000154929.1 |
Stx18
|
syntaxin 18 |
| chr1_+_53313622 | 0.89 |
ENSMUST00000027265.3
|
Osgepl1
|
O-sialoglycoprotein endopeptidase-like 1 |
| chr5_-_74531619 | 0.88 |
ENSMUST00000113542.2
ENSMUST00000072857.6 ENSMUST00000121330.1 ENSMUST00000151474.1 |
Scfd2
|
Sec1 family domain containing 2 |
| chr15_+_65787023 | 0.88 |
ENSMUST00000015146.9
ENSMUST00000173858.1 ENSMUST00000172756.1 ENSMUST00000174856.1 |
Efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr5_-_65335564 | 0.87 |
ENSMUST00000172780.1
|
Rfc1
|
replication factor C (activator 1) 1 |
| chr16_+_22009471 | 0.87 |
ENSMUST00000023561.7
|
Senp2
|
SUMO/sentrin specific peptidase 2 |
| chr10_-_30618337 | 0.87 |
ENSMUST00000019925.5
|
Hint3
|
histidine triad nucleotide binding protein 3 |
| chr7_-_55962424 | 0.87 |
ENSMUST00000126604.1
ENSMUST00000117812.1 ENSMUST00000119201.1 |
Nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 homolog (human) |
| chr1_-_106796609 | 0.85 |
ENSMUST00000112736.1
|
Vps4b
|
vacuolar protein sorting 4b (yeast) |
| chr7_-_55962466 | 0.85 |
ENSMUST00000032635.7
ENSMUST00000152649.1 |
Nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 homolog (human) |
| chr17_-_27907706 | 0.85 |
ENSMUST00000025057.4
|
Taf11
|
TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr9_+_123113215 | 0.85 |
ENSMUST00000026891.4
|
Exosc7
|
exosome component 7 |
| chr14_-_76110760 | 0.85 |
ENSMUST00000022585.3
|
Gpalpp1
|
GPALPP motifs containing 1 |
| chr15_-_81400043 | 0.84 |
ENSMUST00000172107.1
ENSMUST00000169204.1 ENSMUST00000163382.1 |
St13
|
suppression of tumorigenicity 13 |
| chr10_-_30618436 | 0.84 |
ENSMUST00000161074.1
|
Hint3
|
histidine triad nucleotide binding protein 3 |
| chr5_+_48372363 | 0.84 |
ENSMUST00000030968.2
|
Pacrgl
|
PARK2 co-regulated-like |
| chr18_-_35498856 | 0.83 |
ENSMUST00000025215.8
|
Sil1
|
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
| chr10_-_82764088 | 0.83 |
ENSMUST00000130911.1
|
Nfyb
|
nuclear transcription factor-Y beta |
| chr17_+_25184561 | 0.83 |
ENSMUST00000088307.3
|
BC003965
|
cDNA sequence BC003965 |
| chr3_-_100685431 | 0.83 |
ENSMUST00000008907.7
|
Man1a2
|
mannosidase, alpha, class 1A, member 2 |
| chr6_+_57703051 | 0.83 |
ENSMUST00000151042.1
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr19_+_5041337 | 0.82 |
ENSMUST00000116567.2
|
Brms1
|
breast cancer metastasis-suppressor 1 |
| chr5_+_35583018 | 0.82 |
ENSMUST00000068947.7
ENSMUST00000114237.1 ENSMUST00000156125.1 ENSMUST00000068563.5 |
Acox3
|
acyl-Coenzyme A oxidase 3, pristanoyl |
| chr1_+_53313636 | 0.81 |
ENSMUST00000114484.1
|
Osgepl1
|
O-sialoglycoprotein endopeptidase-like 1 |
| chr5_-_36748639 | 0.81 |
ENSMUST00000071949.3
|
Bloc1s4
|
biogenesis of organelles complex-1, subunit 4, cappuccino |
| chr2_-_34826187 | 0.81 |
ENSMUST00000113075.1
ENSMUST00000113080.2 ENSMUST00000091020.3 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr6_+_114643106 | 0.80 |
ENSMUST00000169310.3
ENSMUST00000182169.1 ENSMUST00000183165.1 ENSMUST00000182098.1 ENSMUST00000182793.1 ENSMUST00000182902.1 ENSMUST00000182428.1 ENSMUST00000182035.1 |
Atg7
|
autophagy related 7 |
| chr5_+_67260696 | 0.80 |
ENSMUST00000161233.1
ENSMUST00000160352.1 |
Tmem33
|
transmembrane protein 33 |
| chrX_-_74023908 | 0.80 |
ENSMUST00000033769.8
ENSMUST00000114352.1 ENSMUST00000068286.5 ENSMUST00000114360.3 ENSMUST00000114354.3 |
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr4_+_59003121 | 0.80 |
ENSMUST00000095070.3
ENSMUST00000174664.1 |
Dnajc25
Gm20503
|
DnaJ (Hsp40) homolog, subfamily C, member 25 predicted gene 20503 |
| chr9_+_64179289 | 0.79 |
ENSMUST00000034965.6
|
Snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chrX_-_20962005 | 0.79 |
ENSMUST00000123836.1
|
Uxt
|
ubiquitously expressed transcript |
| chr12_-_98901478 | 0.78 |
ENSMUST00000065716.6
|
Eml5
|
echinoderm microtubule associated protein like 5 |
| chr5_-_76905463 | 0.78 |
ENSMUST00000146570.1
ENSMUST00000142450.1 ENSMUST00000120963.1 |
Aasdh
|
aminoadipate-semialdehyde dehydrogenase |
| chr3_+_138143846 | 0.78 |
ENSMUST00000159481.1
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr6_-_86733268 | 0.78 |
ENSMUST00000001185.7
|
Gmcl1
|
germ cell-less homolog 1 (Drosophila) |
| chr10_-_127041513 | 0.78 |
ENSMUST00000116231.2
|
Mettl21b
|
methyltransferase like 21B |
| chr5_+_124579134 | 0.77 |
ENSMUST00000031333.3
|
Gtf2h3
|
general transcription factor IIH, polypeptide 3 |
| chr10_+_127041931 | 0.77 |
ENSMUST00000006915.7
ENSMUST00000120542.1 |
Mettl1
|
methyltransferase like 1 |
| chr2_+_112454997 | 0.77 |
ENSMUST00000069747.5
|
Emc7
|
ER membrane protein complex subunit 7 |
| chr2_-_34826071 | 0.76 |
ENSMUST00000113077.1
ENSMUST00000028220.3 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr5_+_115341225 | 0.75 |
ENSMUST00000031508.4
|
Triap1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr10_-_81545175 | 0.74 |
ENSMUST00000043604.5
|
Gna11
|
guanine nucleotide binding protein, alpha 11 |
| chr2_+_144556306 | 0.74 |
ENSMUST00000155876.1
ENSMUST00000149697.1 |
Sec23b
|
SEC23B (S. cerevisiae) |
| chr13_-_58402449 | 0.73 |
ENSMUST00000177019.1
|
Hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr2_+_76675265 | 0.73 |
ENSMUST00000111920.1
|
Plekha3
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 3 |
| chr9_+_50768228 | 0.72 |
ENSMUST00000042391.6
|
Fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr4_+_126677630 | 0.72 |
ENSMUST00000030642.2
|
Psmb2
|
proteasome (prosome, macropain) subunit, beta type 2 |
| chr4_-_147904643 | 0.72 |
ENSMUST00000150881.1
|
Mfn2
|
mitofusin 2 |
| chr11_+_83302641 | 0.72 |
ENSMUST00000176430.1
ENSMUST00000065692.7 |
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr11_+_93996082 | 0.71 |
ENSMUST00000041956.7
|
Spag9
|
sperm associated antigen 9 |
| chr7_+_16119868 | 0.71 |
ENSMUST00000006178.4
|
Kptn
|
kaptin |
| chr16_+_32247221 | 0.70 |
ENSMUST00000178573.1
ENSMUST00000023474.3 |
Wdr53
|
WD repeat domain 53 |
| chr10_+_36974536 | 0.70 |
ENSMUST00000019911.7
|
Hdac2
|
histone deacetylase 2 |
| chr16_-_18248697 | 0.70 |
ENSMUST00000115645.3
|
Ranbp1
|
RAN binding protein 1 |
| chr8_+_34154563 | 0.70 |
ENSMUST00000033933.5
|
Tmem66
|
transmembrane protein 66 |
| chr13_-_58215615 | 0.69 |
ENSMUST00000058735.5
ENSMUST00000076454.6 |
Ubqln1
|
ubiquilin 1 |
| chr4_+_98923845 | 0.69 |
ENSMUST00000091358.4
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr17_+_46646225 | 0.69 |
ENSMUST00000002844.7
ENSMUST00000113429.1 ENSMUST00000113430.1 |
Mrpl2
|
mitochondrial ribosomal protein L2 |
| chr5_-_24577467 | 0.68 |
ENSMUST00000030795.8
|
Abcf2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr9_-_50768109 | 0.67 |
ENSMUST00000177546.1
ENSMUST00000176238.1 |
1110032A03Rik
|
RIKEN cDNA 1110032A03 gene |
| chr19_+_3282901 | 0.67 |
ENSMUST00000025745.3
ENSMUST00000025743.6 |
Mrpl21
|
mitochondrial ribosomal protein L21 |
| chr13_+_73604002 | 0.67 |
ENSMUST00000022102.7
|
Clptm1l
|
CLPTM1-like |
| chr1_-_59120079 | 0.67 |
ENSMUST00000094917.3
|
Tmem237
|
transmembrane protein 237 |
| chr1_-_59119748 | 0.66 |
ENSMUST00000087475.4
|
Tmem237
|
transmembrane protein 237 |
| chr7_+_138846579 | 0.66 |
ENSMUST00000155672.1
|
Ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta isoform |
| chr17_+_14978864 | 0.66 |
ENSMUST00000061688.9
|
9030025P20Rik
|
RIKEN cDNA 9030025P20 gene |
| chr10_+_58446845 | 0.65 |
ENSMUST00000003310.5
|
Ranbp2
|
RAN binding protein 2 |
| chrX_+_57053549 | 0.65 |
ENSMUST00000114751.2
ENSMUST00000088652.5 |
Htatsf1
|
HIV TAT specific factor 1 |
| chr3_+_138143888 | 0.63 |
ENSMUST00000161141.1
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr11_+_60417238 | 0.63 |
ENSMUST00000070681.6
|
Gid4
|
GID complex subunit 4, VID24 homolog (S. cerevisiae) |
| chr2_-_103761178 | 0.63 |
ENSMUST00000028608.6
|
Nat10
|
N-acetyltransferase 10 |
| chr9_-_44965519 | 0.62 |
ENSMUST00000125642.1
ENSMUST00000117506.1 ENSMUST00000117549.1 |
Ube4a
|
ubiquitination factor E4A, UFD2 homolog (S. cerevisiae) |
| chr11_-_98587193 | 0.62 |
ENSMUST00000052919.7
|
Ormdl3
|
ORM1-like 3 (S. cerevisiae) |
| chr3_+_103058302 | 0.61 |
ENSMUST00000029445.6
|
Nras
|
neuroblastoma ras oncogene |
| chr7_-_35056467 | 0.61 |
ENSMUST00000130491.1
|
Cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr5_-_124579045 | 0.60 |
ENSMUST00000135361.1
ENSMUST00000031334.8 |
Eif2b1
|
eukaryotic translation initiation factor 2B, subunit 1 (alpha) |
| chr10_+_80141457 | 0.59 |
ENSMUST00000105367.1
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr7_+_132931142 | 0.59 |
ENSMUST00000106157.1
|
Zranb1
|
zinc finger, RAN-binding domain containing 1 |
| chr4_+_98923810 | 0.58 |
ENSMUST00000030289.2
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr5_-_115341142 | 0.57 |
ENSMUST00000139167.1
|
Gatc
|
glutamyl-tRNA(Gln) amidotransferase, subunit C homolog (bacterial) |
| chr16_-_4077778 | 0.57 |
ENSMUST00000006137.8
|
Trap1
|
TNF receptor-associated protein 1 |
| chr8_+_117157972 | 0.57 |
ENSMUST00000064488.4
ENSMUST00000162997.1 |
Gan
|
giant axonal neuropathy |
| chr16_+_14705832 | 0.56 |
ENSMUST00000023356.6
|
Snai2
|
snail homolog 2 (Drosophila) |
| chr11_+_70700473 | 0.56 |
ENSMUST00000152618.2
ENSMUST00000102554.1 ENSMUST00000094499.4 ENSMUST00000072187.5 |
Kif1c
|
kinesin family member 1C |
| chr11_+_96034885 | 0.55 |
ENSMUST00000006217.3
ENSMUST00000107700.3 |
Snf8
|
SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) |
| chr11_-_76243687 | 0.55 |
ENSMUST00000017430.5
|
Glod4
|
glyoxalase domain containing 4 |
| chr5_+_29735940 | 0.55 |
ENSMUST00000114839.1
|
Dnajb6
|
DnaJ (Hsp40) homolog, subfamily B, member 6 |
| chr13_-_58402476 | 0.54 |
ENSMUST00000175847.1
ENSMUST00000043269.7 ENSMUST00000177060.1 |
Hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr2_+_152226839 | 0.54 |
ENSMUST00000099224.3
ENSMUST00000124791.1 ENSMUST00000133119.1 |
Csnk2a1
|
casein kinase 2, alpha 1 polypeptide |
| chr17_-_27820445 | 0.54 |
ENSMUST00000114859.1
|
D17Wsu92e
|
DNA segment, Chr 17, Wayne State University 92, expressed |
| chrX_-_36864238 | 0.54 |
ENSMUST00000115249.3
ENSMUST00000115248.3 |
C330007P06Rik
|
RIKEN cDNA C330007P06 gene |
| chr19_-_6057736 | 0.54 |
ENSMUST00000007482.6
|
Mrpl49
|
mitochondrial ribosomal protein L49 |
| chr6_+_114643094 | 0.53 |
ENSMUST00000032457.10
|
Atg7
|
autophagy related 7 |
| chr8_+_27023793 | 0.53 |
ENSMUST00000033873.7
|
Erlin2
|
ER lipid raft associated 2 |
| chr3_+_36552600 | 0.52 |
ENSMUST00000029269.5
ENSMUST00000136890.1 |
Exosc9
|
exosome component 9 |
| chr4_+_107879745 | 0.52 |
ENSMUST00000030348.5
|
Magoh
|
mago-nashi homolog, proliferation-associated (Drosophila) |
| chr12_+_52097737 | 0.52 |
ENSMUST00000040090.9
|
Nubpl
|
nucleotide binding protein-like |
| chr18_-_68300194 | 0.51 |
ENSMUST00000152193.1
|
Fam210a
|
family with sequence similarity 210, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Creb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.7 | 2.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.7 | 2.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.6 | 1.9 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.6 | 1.8 | GO:1901950 | dense core granule localization(GO:0032253) dense core granule transport(GO:1901950) regulation of dense core granule transport(GO:1904809) positive regulation of dense core granule transport(GO:1904811) |
| 0.6 | 1.7 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.5 | 5.1 | GO:0070236 | phospholipid homeostasis(GO:0055091) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.5 | 1.5 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.5 | 2.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.5 | 1.9 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.5 | 5.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.5 | 2.3 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.5 | 1.4 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.4 | 1.3 | GO:0090157 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.4 | 1.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.4 | 1.7 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.4 | 1.3 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.4 | 1.1 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.3 | 1.4 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.3 | 1.3 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.3 | 2.9 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.3 | 0.3 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.3 | 1.6 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.3 | 1.2 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.3 | 0.9 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.3 | 1.2 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.3 | 1.9 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.3 | 1.1 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.2 | 1.5 | GO:0060701 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.2 | 1.7 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.2 | 1.0 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.2 | 0.7 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.2 | 1.9 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.2 | 1.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.2 | 4.0 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.2 | 2.5 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 0.6 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.2 | 2.8 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.2 | 3.2 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.2 | 0.6 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.2 | 0.6 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.2 | 0.6 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.2 | 2.6 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 | 3.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 0.7 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 1.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.2 | 0.3 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.2 | 0.5 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.2 | 0.7 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.2 | 0.8 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.2 | 1.8 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.2 | 0.5 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 1.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 1.5 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.2 | 0.9 | GO:0051189 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.2 | 1.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 1.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.4 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 0.8 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 1.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.8 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.8 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.5 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.4 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.1 | 0.5 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.7 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 1.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 0.7 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) regulation of zinc ion transport(GO:0071579) |
| 0.1 | 3.1 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 1.2 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.3 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.1 | 1.4 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 1.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.4 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 2.0 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 1.0 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.9 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.5 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.6 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 1.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.8 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.1 | 0.9 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 3.8 | GO:0043039 | tRNA aminoacylation(GO:0043039) |
| 0.1 | 0.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.3 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.4 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 0.5 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.9 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.6 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.3 | GO:0051309 | female meiosis chromosome separation(GO:0051309) |
| 0.1 | 0.2 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 0.7 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 2.6 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.4 | GO:1903182 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.1 | 1.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.2 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.3 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.4 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 1.2 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 0.4 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 2.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.2 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.8 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.5 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.4 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.4 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 1.0 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.4 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 1.2 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.5 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.1 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 1.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.5 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.1 | GO:0035188 | blastocyst hatching(GO:0001835) DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 1.2 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 4.7 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 2.7 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.4 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.0 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.4 | GO:0051452 | intracellular pH reduction(GO:0051452) |
| 0.0 | 0.3 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.0 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.6 | 3.6 | GO:1990393 | 3M complex(GO:1990393) |
| 0.5 | 1.5 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.5 | 2.0 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.5 | 2.7 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.5 | 3.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.3 | 1.7 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.3 | 2.0 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.3 | 2.4 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.2 | 0.7 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.2 | 2.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 3.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 0.6 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.2 | 2.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 1.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 0.6 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 7.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.8 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.9 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 1.9 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.6 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 1.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 1.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.5 | GO:0032444 | activin responsive factor complex(GO:0032444) SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.8 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 1.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.4 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 1.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.4 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.6 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 2.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 1.0 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 2.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.4 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.1 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 3.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 1.7 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.1 | 1.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.4 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 9.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 0.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 1.8 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.8 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.7 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 1.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.7 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.8 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 1.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 5.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 3.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.5 | 4.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.5 | 1.5 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.4 | 1.3 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.4 | 1.6 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.4 | 1.2 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.4 | 2.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.4 | 2.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.3 | 1.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.3 | 2.5 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.3 | 1.2 | GO:0035851 | histone deacetylase activity (H4-K16 specific)(GO:0034739) Krueppel-associated box domain binding(GO:0035851) |
| 0.3 | 0.9 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.3 | 0.8 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 1.5 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.2 | 0.7 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 2.0 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.2 | 1.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 1.9 | GO:0032564 | dATP binding(GO:0032564) |
| 0.2 | 0.6 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.2 | 3.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 2.0 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 5.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 1.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 3.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 1.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.4 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 0.7 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.8 | GO:0004043 | L-aminoadipate-semialdehyde dehydrogenase activity(GO:0004043) |
| 0.1 | 2.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 3.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.5 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 0.3 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.5 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 2.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.4 | GO:0016433 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.1 | 2.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 5.5 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.1 | 0.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 1.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.3 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 3.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 1.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.3 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 0.8 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.4 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.8 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.9 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 2.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.4 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 0.4 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.4 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 5.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 2.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 1.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.5 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 0.9 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 1.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.7 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 1.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 1.0 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.6 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 1.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 3.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.9 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 2.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 3.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.4 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.6 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 1.3 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 1.1 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.3 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 7.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 3.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 4.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 2.6 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 2.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 1.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 3.6 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 3.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 7.4 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 2.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 0.3 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.1 | 1.5 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.1 | 0.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.6 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 2.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 2.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 4.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |