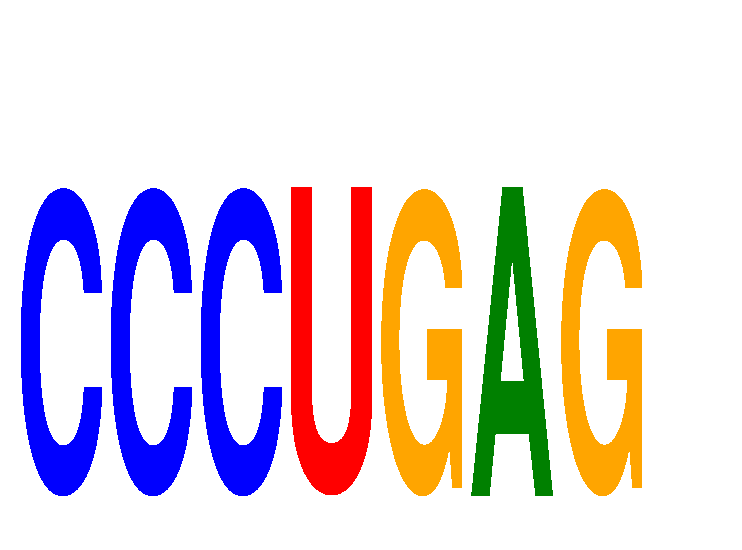Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for CCCUGAG
Z-value: 0.68
miRNA associated with seed CCCUGAG
| Name | miRBASE accession |
|---|---|
|
mmu-miR-125a-5p
|
MIMAT0000135 |
|
mmu-miR-125b-5p
|
MIMAT0000136 |
|
mmu-miR-351-5p
|
MIMAT0000609 |
|
mmu-miR-6367
|
MIMAT0025111 |
|
mmu-miR-6394
|
MIMAT0025144 |
Activity profile of CCCUGAG motif
Sorted Z-values of CCCUGAG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_12037391 | 2.14 |
ENSMUST00000093321.5
|
Grb10
|
growth factor receptor bound protein 10 |
| chr4_+_136172367 | 1.66 |
ENSMUST00000061721.5
|
E2f2
|
E2F transcription factor 2 |
| chr4_-_134018829 | 1.54 |
ENSMUST00000051674.2
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr12_+_24831583 | 1.51 |
ENSMUST00000110942.3
ENSMUST00000078902.6 |
Mboat2
|
membrane bound O-acyltransferase domain containing 2 |
| chr2_-_24763047 | 1.42 |
ENSMUST00000100348.3
ENSMUST00000041342.5 ENSMUST00000114447.1 ENSMUST00000102939.2 ENSMUST00000070864.7 |
Cacna1b
|
calcium channel, voltage-dependent, N type, alpha 1B subunit |
| chr19_-_17356631 | 1.29 |
ENSMUST00000174236.1
|
Gcnt1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr10_+_79927039 | 1.26 |
ENSMUST00000019708.5
ENSMUST00000105377.1 |
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr11_+_95009852 | 1.21 |
ENSMUST00000055947.3
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr10_-_45470201 | 1.20 |
ENSMUST00000079390.6
|
Lin28b
|
lin-28 homolog B (C. elegans) |
| chr15_-_73184840 | 1.18 |
ENSMUST00000044113.10
|
Ago2
|
argonaute RISC catalytic subunit 2 |
| chr18_+_61953048 | 1.14 |
ENSMUST00000051720.5
|
Sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr5_-_148399901 | 1.10 |
ENSMUST00000048116.8
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr5_-_30945393 | 0.98 |
ENSMUST00000031051.6
|
Cgref1
|
cell growth regulator with EF hand domain 1 |
| chr13_+_30749226 | 0.96 |
ENSMUST00000021784.2
ENSMUST00000110307.1 |
Irf4
|
interferon regulatory factor 4 |
| chr8_+_79028587 | 0.93 |
ENSMUST00000119254.1
|
Zfp827
|
zinc finger protein 827 |
| chr4_-_46566432 | 0.92 |
ENSMUST00000030021.7
ENSMUST00000107757.1 |
Coro2a
|
coronin, actin binding protein 2A |
| chr9_-_108190352 | 0.91 |
ENSMUST00000035208.7
|
Bsn
|
bassoon |
| chr14_+_70457447 | 0.84 |
ENSMUST00000003561.3
|
Phyhip
|
phytanoyl-CoA hydroxylase interacting protein |
| chr8_-_31918203 | 0.80 |
ENSMUST00000073884.4
|
Nrg1
|
neuregulin 1 |
| chr13_-_51793650 | 0.80 |
ENSMUST00000110040.2
ENSMUST00000021900.7 |
Sema4d
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr9_+_45138437 | 0.77 |
ENSMUST00000060125.5
|
Scn4b
|
sodium channel, type IV, beta |
| chr4_-_45084538 | 0.76 |
ENSMUST00000052236.6
|
Fbxo10
|
F-box protein 10 |
| chr2_+_130295148 | 0.73 |
ENSMUST00000110288.2
|
Ebf4
|
early B cell factor 4 |
| chr11_+_116198853 | 0.73 |
ENSMUST00000021130.6
|
Ten1
|
TEN1 telomerase capping complex subunit |
| chr4_-_133887765 | 0.72 |
ENSMUST00000003741.9
ENSMUST00000105894.4 |
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr11_+_43528759 | 0.71 |
ENSMUST00000050574.6
|
Ccnjl
|
cyclin J-like |
| chr5_+_123076275 | 0.70 |
ENSMUST00000067505.8
ENSMUST00000111619.3 ENSMUST00000160344.1 |
Tmem120b
|
transmembrane protein 120B |
| chr9_+_72806874 | 0.66 |
ENSMUST00000055535.8
|
Prtg
|
protogenin homolog (Gallus gallus) |
| chr11_+_94990996 | 0.66 |
ENSMUST00000038696.5
|
Ppp1r9b
|
protein phosphatase 1, regulatory subunit 9B |
| chr16_-_20426375 | 0.64 |
ENSMUST00000079158.6
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr2_-_154558834 | 0.64 |
ENSMUST00000109716.2
ENSMUST00000000895.6 ENSMUST00000125793.1 |
Necab3
|
N-terminal EF-hand calcium binding protein 3 |
| chr11_-_102819114 | 0.63 |
ENSMUST00000068933.5
|
Gjc1
|
gap junction protein, gamma 1 |
| chr10_-_80577285 | 0.62 |
ENSMUST00000038558.8
|
Klf16
|
Kruppel-like factor 16 |
| chr14_+_116925291 | 0.61 |
ENSMUST00000078849.4
|
Gpc6
|
glypican 6 |
| chr5_-_38561658 | 0.59 |
ENSMUST00000005234.9
|
Wdr1
|
WD repeat domain 1 |
| chr10_-_128401218 | 0.58 |
ENSMUST00000042666.5
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr7_+_4740111 | 0.57 |
ENSMUST00000098853.2
|
Suv420h2
|
suppressor of variegation 4-20 homolog 2 (Drosophila) |
| chr12_-_81781121 | 0.57 |
ENSMUST00000035987.7
|
Map3k9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr6_+_42264983 | 0.56 |
ENSMUST00000031895.6
|
Casp2
|
caspase 2 |
| chr8_+_106168857 | 0.56 |
ENSMUST00000034378.3
|
Slc7a6
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 |
| chr7_+_27258725 | 0.55 |
ENSMUST00000079258.6
|
Numbl
|
numb-like |
| chr11_-_106160101 | 0.55 |
ENSMUST00000045923.3
|
Limd2
|
LIM domain containing 2 |
| chr17_+_28142267 | 0.54 |
ENSMUST00000043503.3
|
Scube3
|
signal peptide, CUB domain, EGF-like 3 |
| chr11_-_4594750 | 0.54 |
ENSMUST00000109943.3
|
Mtmr3
|
myotubularin related protein 3 |
| chr7_-_83735503 | 0.53 |
ENSMUST00000001792.4
|
Il16
|
interleukin 16 |
| chr3_+_95228732 | 0.52 |
ENSMUST00000053872.5
|
Cdc42se1
|
CDC42 small effector 1 |
| chr9_+_62838767 | 0.51 |
ENSMUST00000034776.6
|
Cln6
|
ceroid-lipofuscinosis, neuronal 6 |
| chr11_+_7063423 | 0.51 |
ENSMUST00000020706.4
|
Adcy1
|
adenylate cyclase 1 |
| chr5_+_140607334 | 0.50 |
ENSMUST00000031555.1
|
Lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr18_+_10325148 | 0.49 |
ENSMUST00000048977.8
|
Greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr14_+_60378242 | 0.49 |
ENSMUST00000022561.6
|
Amer2
|
APC membrane recruitment 2 |
| chr2_+_32876114 | 0.49 |
ENSMUST00000028135.8
|
Fam129b
|
family with sequence similarity 129, member B |
| chr6_-_91116785 | 0.49 |
ENSMUST00000113509.1
ENSMUST00000032179.7 |
Nup210
|
nucleoporin 210 |
| chr10_+_42860348 | 0.49 |
ENSMUST00000063063.7
|
Scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr7_+_126272589 | 0.49 |
ENSMUST00000056028.9
|
Sbk1
|
SH3-binding kinase 1 |
| chr6_-_42324640 | 0.48 |
ENSMUST00000031891.8
ENSMUST00000143278.1 |
Fam131b
|
family with sequence similarity 131, member B |
| chr7_-_122021143 | 0.48 |
ENSMUST00000033160.8
|
Gga2
|
golgi associated, gamma adaptin ear containing, ARF binding protein 2 |
| chr12_-_27342696 | 0.48 |
ENSMUST00000079063.5
|
Sox11
|
SRY-box containing gene 11 |
| chr12_-_78983476 | 0.47 |
ENSMUST00000070174.7
|
Tmem229b
|
transmembrane protein 229B |
| chr11_+_98836775 | 0.47 |
ENSMUST00000107479.2
|
Rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr10_-_128525859 | 0.46 |
ENSMUST00000026427.6
|
Esyt1
|
extended synaptotagmin-like protein 1 |
| chr10_-_61273409 | 0.46 |
ENSMUST00000092486.4
|
Adamts14
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 14 |
| chr9_-_66919646 | 0.45 |
ENSMUST00000041139.7
|
Rab8b
|
RAB8B, member RAS oncogene family |
| chr6_-_31563978 | 0.45 |
ENSMUST00000026698.7
|
Podxl
|
podocalyxin-like |
| chr12_+_32378692 | 0.45 |
ENSMUST00000172332.2
|
Ccdc71l
|
coiled-coil domain containing 71 like |
| chr15_-_99457742 | 0.45 |
ENSMUST00000023747.7
|
Nckap5l
|
NCK-associated protein 5-like |
| chr2_-_25608447 | 0.44 |
ENSMUST00000058137.8
|
Rabl6
|
RAB, member of RAS oncogene family-like 6 |
| chr2_-_151009364 | 0.44 |
ENSMUST00000109896.1
|
Ninl
|
ninein-like |
| chr17_+_35861318 | 0.43 |
ENSMUST00000074259.8
ENSMUST00000174873.1 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr6_+_118066356 | 0.43 |
ENSMUST00000164960.1
|
Rasgef1a
|
RasGEF domain family, member 1A |
| chr2_+_37776229 | 0.43 |
ENSMUST00000050372.7
|
Crb2
|
crumbs homolog 2 (Drosophila) |
| chr9_+_119937606 | 0.43 |
ENSMUST00000035100.5
|
Ttc21a
|
tetratricopeptide repeat domain 21A |
| chr7_+_16130285 | 0.42 |
ENSMUST00000168693.1
|
Slc8a2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr9_-_30922452 | 0.42 |
ENSMUST00000065112.6
|
Adamts15
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
| chr2_-_33431324 | 0.42 |
ENSMUST00000113158.1
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chr2_+_167932327 | 0.42 |
ENSMUST00000029053.7
|
Ptpn1
|
protein tyrosine phosphatase, non-receptor type 1 |
| chr9_+_21265287 | 0.41 |
ENSMUST00000065005.3
|
Atg4d
|
autophagy related 4D, cysteine peptidase |
| chr5_+_115429585 | 0.41 |
ENSMUST00000150779.1
|
Msi1
|
musashi RNA-binding protein 1 |
| chr10_-_23787195 | 0.40 |
ENSMUST00000073926.6
|
Rps12
|
ribosomal protein S12 |
| chr1_-_167285110 | 0.40 |
ENSMUST00000027839.8
|
Uck2
|
uridine-cytidine kinase 2 |
| chr15_+_99224976 | 0.40 |
ENSMUST00000041415.3
|
Kcnh3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr10_-_128891674 | 0.40 |
ENSMUST00000026408.6
|
Gdf11
|
growth differentiation factor 11 |
| chr4_+_141147911 | 0.40 |
ENSMUST00000030757.9
|
Fbxo42
|
F-box protein 42 |
| chr16_-_58523349 | 0.39 |
ENSMUST00000137035.1
|
St3gal6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr17_-_35188427 | 0.39 |
ENSMUST00000097336.4
|
Lst1
|
leukocyte specific transcript 1 |
| chr3_+_28781305 | 0.39 |
ENSMUST00000060500.7
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr7_+_27473761 | 0.38 |
ENSMUST00000068641.6
|
Sertad3
|
SERTA domain containing 3 |
| chr15_-_50889691 | 0.38 |
ENSMUST00000165201.2
ENSMUST00000184458.1 |
Trps1
|
trichorhinophalangeal syndrome I (human) |
| chr3_-_9610074 | 0.38 |
ENSMUST00000041124.7
|
Zfp704
|
zinc finger protein 704 |
| chr1_-_184999549 | 0.38 |
ENSMUST00000027929.4
|
Mark1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr9_-_62537036 | 0.38 |
ENSMUST00000048043.5
|
Coro2b
|
coronin, actin binding protein, 2B |
| chr17_-_27029009 | 0.38 |
ENSMUST00000078691.5
|
Bak1
|
BCL2-antagonist/killer 1 |
| chr1_-_52952834 | 0.37 |
ENSMUST00000050567.4
|
1700019D03Rik
|
RIKEN cDNA 1700019D03 gene |
| chr19_-_5912834 | 0.37 |
ENSMUST00000136983.1
|
Dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr19_+_41482632 | 0.37 |
ENSMUST00000067795.5
|
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr19_-_10457447 | 0.36 |
ENSMUST00000171400.2
|
Lrrc10b
|
leucine rich repeat containing 10B |
| chr10_+_117629493 | 0.36 |
ENSMUST00000020399.5
|
Cpm
|
carboxypeptidase M |
| chr7_+_98835104 | 0.36 |
ENSMUST00000165122.1
ENSMUST00000067495.2 |
Wnt11
|
wingless-related MMTV integration site 11 |
| chr4_+_86874396 | 0.36 |
ENSMUST00000045224.7
ENSMUST00000084433.4 |
Acer2
|
alkaline ceramidase 2 |
| chr14_-_110755100 | 0.35 |
ENSMUST00000078386.2
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr18_+_22345089 | 0.34 |
ENSMUST00000120223.1
ENSMUST00000097655.3 |
Asxl3
|
additional sex combs like 3 (Drosophila) |
| chr18_-_20746402 | 0.34 |
ENSMUST00000070080.5
|
B4galt6
|
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 6 |
| chr7_+_97081711 | 0.34 |
ENSMUST00000004622.5
|
Gab2
|
growth factor receptor bound protein 2-associated protein 2 |
| chr7_+_45617575 | 0.33 |
ENSMUST00000008605.5
|
Fut1
|
fucosyltransferase 1 |
| chr1_-_134079114 | 0.33 |
ENSMUST00000020692.6
|
Btg2
|
B cell translocation gene 2, anti-proliferative |
| chr2_+_25242929 | 0.33 |
ENSMUST00000114355.1
ENSMUST00000060818.1 |
Rnf208
|
ring finger protein 208 |
| chrX_-_8074720 | 0.33 |
ENSMUST00000115636.3
ENSMUST00000115638.3 |
Suv39h1
|
suppressor of variegation 3-9 homolog 1 (Drosophila) |
| chr15_-_89140664 | 0.33 |
ENSMUST00000088827.6
|
Mapk12
|
mitogen-activated protein kinase 12 |
| chr17_+_46161021 | 0.33 |
ENSMUST00000024748.7
ENSMUST00000172170.1 |
Gtpbp2
|
GTP binding protein 2 |
| chr1_-_95667555 | 0.33 |
ENSMUST00000043336.4
|
St8sia4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr4_+_128654686 | 0.33 |
ENSMUST00000030588.6
ENSMUST00000136377.1 |
Phc2
|
polyhomeotic-like 2 (Drosophila) |
| chr16_+_5050012 | 0.32 |
ENSMUST00000052449.5
|
Ubn1
|
ubinuclein 1 |
| chr8_+_4238733 | 0.32 |
ENSMUST00000110998.2
ENSMUST00000062686.4 |
Map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr10_-_80671976 | 0.32 |
ENSMUST00000003433.6
|
Mknk2
|
MAP kinase-interacting serine/threonine kinase 2 |
| chr15_-_97908261 | 0.32 |
ENSMUST00000023119.8
|
Vdr
|
vitamin D receptor |
| chr8_+_83608175 | 0.32 |
ENSMUST00000005620.8
|
Dnajb1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr2_-_153241402 | 0.31 |
ENSMUST00000056924.7
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr5_-_124032214 | 0.31 |
ENSMUST00000040967.7
|
Vps37b
|
vacuolar protein sorting 37B (yeast) |
| chr17_-_29888570 | 0.31 |
ENSMUST00000171691.1
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr2_-_118549668 | 0.31 |
ENSMUST00000090219.6
|
Bmf
|
BCL2 modifying factor |
| chr3_-_19311269 | 0.31 |
ENSMUST00000099195.3
|
Pde7a
|
phosphodiesterase 7A |
| chr3_-_27710413 | 0.31 |
ENSMUST00000046157.4
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr6_-_88841935 | 0.31 |
ENSMUST00000032169.5
|
Abtb1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr19_-_5085483 | 0.30 |
ENSMUST00000140389.1
ENSMUST00000151413.1 ENSMUST00000077066.7 |
Tmem151a
|
transmembrane protein 151A |
| chr19_-_7241216 | 0.30 |
ENSMUST00000025675.9
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr8_-_124021309 | 0.30 |
ENSMUST00000165628.1
|
Taf5l
|
TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr1_+_89070406 | 0.30 |
ENSMUST00000066279.4
|
Sh3bp4
|
SH3-domain binding protein 4 |
| chr10_+_126978690 | 0.30 |
ENSMUST00000105256.2
|
Ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr1_-_58504946 | 0.30 |
ENSMUST00000027198.5
|
Orc2
|
origin recognition complex, subunit 2 |
| chr17_+_28801090 | 0.29 |
ENSMUST00000004985.9
|
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr1_-_36558349 | 0.29 |
ENSMUST00000114991.1
ENSMUST00000114992.1 |
Sema4c
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr2_-_181599128 | 0.29 |
ENSMUST00000060173.8
|
Samd10
|
sterile alpha motif domain containing 10 |
| chr7_+_130577334 | 0.29 |
ENSMUST00000059145.7
ENSMUST00000084513.4 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr17_-_46282991 | 0.29 |
ENSMUST00000180283.1
ENSMUST00000012440.6 ENSMUST00000164342.2 |
Tjap1
|
tight junction associated protein 1 |
| chr8_-_120101473 | 0.29 |
ENSMUST00000034280.7
|
Zdhhc7
|
zinc finger, DHHC domain containing 7 |
| chr11_-_98545800 | 0.29 |
ENSMUST00000103141.3
|
Ikzf3
|
IKAROS family zinc finger 3 |
| chr16_+_17233560 | 0.29 |
ENSMUST00000090190.5
ENSMUST00000115698.2 |
Hic2
|
hypermethylated in cancer 2 |
| chr2_-_155514796 | 0.29 |
ENSMUST00000029131.4
|
Ggt7
|
gamma-glutamyltransferase 7 |
| chr1_-_153549697 | 0.28 |
ENSMUST00000041874.7
|
Npl
|
N-acetylneuraminate pyruvate lyase |
| chr1_-_37541003 | 0.28 |
ENSMUST00000151952.1
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr11_-_60913775 | 0.28 |
ENSMUST00000019075.3
|
Gm16515
|
predicted gene, Gm16515 |
| chr6_-_82774448 | 0.27 |
ENSMUST00000000642.4
|
Hk2
|
hexokinase 2 |
| chr10_-_19015347 | 0.27 |
ENSMUST00000019997.4
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr5_-_113650390 | 0.27 |
ENSMUST00000047936.6
|
Cmklr1
|
chemokine-like receptor 1 |
| chr11_+_52098681 | 0.26 |
ENSMUST00000020608.2
|
Ppp2ca
|
protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform |
| chr8_-_70439557 | 0.26 |
ENSMUST00000076615.5
|
Crtc1
|
CREB regulated transcription coactivator 1 |
| chr5_+_91517615 | 0.26 |
ENSMUST00000040576.9
|
Parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chr11_-_46166397 | 0.26 |
ENSMUST00000020679.2
|
Nipal4
|
NIPA-like domain containing 4 |
| chr14_+_69029289 | 0.26 |
ENSMUST00000014957.8
|
Stc1
|
stanniocalcin 1 |
| chr7_-_63938862 | 0.25 |
ENSMUST00000063694.8
|
Klf13
|
Kruppel-like factor 13 |
| chr9_+_108002501 | 0.25 |
ENSMUST00000035214.4
ENSMUST00000175874.1 |
Ip6k1
|
inositol hexaphosphate kinase 1 |
| chr9_-_110654161 | 0.25 |
ENSMUST00000133191.1
ENSMUST00000167320.1 |
Nbeal2
|
neurobeachin-like 2 |
| chr13_+_46418266 | 0.25 |
ENSMUST00000037923.3
|
Rbm24
|
RNA binding motif protein 24 |
| chr12_-_15816762 | 0.25 |
ENSMUST00000020922.7
|
Trib2
|
tribbles homolog 2 (Drosophila) |
| chr11_-_109473598 | 0.24 |
ENSMUST00000070152.5
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr9_-_85327110 | 0.24 |
ENSMUST00000034802.8
|
Fam46a
|
family with sequence similarity 46, member A |
| chr3_+_141465564 | 0.24 |
ENSMUST00000106236.2
ENSMUST00000075282.3 |
Unc5c
|
unc-5 homolog C (C. elegans) |
| chrX_+_106920618 | 0.24 |
ENSMUST00000060576.7
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr11_+_96323253 | 0.23 |
ENSMUST00000093944.3
|
Hoxb3
|
homeobox B3 |
| chr13_-_111808938 | 0.23 |
ENSMUST00000109267.2
|
Map3k1
|
mitogen-activated protein kinase kinase kinase 1 |
| chr10_+_60346851 | 0.23 |
ENSMUST00000020301.7
ENSMUST00000105460.1 ENSMUST00000170507.1 |
4632428N05Rik
|
RIKEN cDNA 4632428N05 gene |
| chr6_-_89362581 | 0.22 |
ENSMUST00000163139.1
|
Plxna1
|
plexin A1 |
| chr7_-_100371889 | 0.22 |
ENSMUST00000032963.8
|
Ppme1
|
protein phosphatase methylesterase 1 |
| chr11_+_3330401 | 0.22 |
ENSMUST00000045153.4
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr3_+_88336256 | 0.21 |
ENSMUST00000001451.5
|
Smg5
|
Smg-5 homolog, nonsense mediated mRNA decay factor (C. elegans) |
| chr4_-_82505749 | 0.21 |
ENSMUST00000107245.2
ENSMUST00000107246.1 |
Nfib
|
nuclear factor I/B |
| chr4_+_152274191 | 0.21 |
ENSMUST00000105650.1
ENSMUST00000105651.1 |
Gpr153
|
G protein-coupled receptor 153 |
| chr18_+_57354733 | 0.21 |
ENSMUST00000025490.8
|
Prrc1
|
proline-rich coiled-coil 1 |
| chr19_+_44562841 | 0.21 |
ENSMUST00000040455.4
|
Hif1an
|
hypoxia-inducible factor 1, alpha subunit inhibitor |
| chr1_+_75142775 | 0.21 |
ENSMUST00000097694.4
|
Fam134a
|
family with sequence similarity 134, member A |
| chr7_+_80186835 | 0.21 |
ENSMUST00000107383.1
ENSMUST00000032754.7 |
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr14_+_31251454 | 0.21 |
ENSMUST00000022458.4
|
Bap1
|
Brca1 associated protein 1 |
| chr18_+_36664060 | 0.21 |
ENSMUST00000036765.7
|
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr3_+_95658714 | 0.21 |
ENSMUST00000037947.8
|
Mcl1
|
myeloid cell leukemia sequence 1 |
| chr8_-_106893581 | 0.21 |
ENSMUST00000176437.1
ENSMUST00000177068.1 ENSMUST00000169312.1 ENSMUST00000176515.1 |
Chtf8
|
CTF8, chromosome transmission fidelity factor 8 |
| chr8_-_109693235 | 0.20 |
ENSMUST00000034164.4
|
Ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr11_-_76509419 | 0.20 |
ENSMUST00000094012.4
|
Abr
|
active BCR-related gene |
| chr1_+_75479529 | 0.20 |
ENSMUST00000113575.2
ENSMUST00000148980.1 ENSMUST00000050899.6 |
Tmem198
|
transmembrane protein 198 |
| chr14_-_118706180 | 0.19 |
ENSMUST00000036554.6
ENSMUST00000166646.1 |
Abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr19_-_10304867 | 0.19 |
ENSMUST00000039327.4
|
Dagla
|
diacylglycerol lipase, alpha |
| chr6_+_83165920 | 0.19 |
ENSMUST00000077407.5
ENSMUST00000113913.1 ENSMUST00000130212.1 |
Dctn1
|
dynactin 1 |
| chr7_+_45163915 | 0.19 |
ENSMUST00000085374.5
|
Slc17a7
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 |
| chr19_+_4192129 | 0.19 |
ENSMUST00000046094.4
|
Ppp1ca
|
protein phosphatase 1, catalytic subunit, alpha isoform |
| chr10_+_82629803 | 0.18 |
ENSMUST00000092266.4
ENSMUST00000151390.1 |
Tdg
|
thymine DNA glycosylase |
| chr11_-_98022594 | 0.18 |
ENSMUST00000103144.3
ENSMUST00000017552.6 ENSMUST00000092736.4 ENSMUST00000107562.1 |
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr6_-_72439549 | 0.18 |
ENSMUST00000059472.8
|
Mat2a
|
methionine adenosyltransferase II, alpha |
| chr9_-_44305595 | 0.18 |
ENSMUST00000034629.4
|
Hinfp
|
histone H4 transcription factor |
| chr14_-_40966807 | 0.18 |
ENSMUST00000047652.5
|
Tspan14
|
tetraspanin 14 |
| chr6_-_53820764 | 0.18 |
ENSMUST00000127748.2
|
Tril
|
TLR4 interactor with leucine-rich repeats |
| chrX_-_137038265 | 0.18 |
ENSMUST00000113070.2
ENSMUST00000113069.2 |
Slc25a53
|
solute carrier family 25, member 53 |
| chr11_+_58330712 | 0.18 |
ENSMUST00000116376.2
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr9_-_21918089 | 0.17 |
ENSMUST00000128442.1
ENSMUST00000119055.1 ENSMUST00000122211.1 ENSMUST00000115351.3 |
Rab3d
|
RAB3D, member RAS oncogene family |
| chr13_+_31625802 | 0.17 |
ENSMUST00000042054.2
|
Foxf2
|
forkhead box F2 |
| chr1_+_167001417 | 0.17 |
ENSMUST00000165874.1
|
Fam78b
|
family with sequence similarity 78, member B |
| chr7_+_99381495 | 0.17 |
ENSMUST00000037528.8
|
Gdpd5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr6_-_83441674 | 0.17 |
ENSMUST00000089622.4
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr12_-_108894116 | 0.17 |
ENSMUST00000109848.3
|
Wars
|
tryptophanyl-tRNA synthetase |
| chr2_+_25332729 | 0.17 |
ENSMUST00000042390.4
|
Man1b1
|
mannosidase, alpha, class 1B, member 1 |
| chr11_+_115814724 | 0.17 |
ENSMUST00000106481.2
ENSMUST00000021134.3 |
Tsen54
|
tRNA splicing endonuclease 54 homolog (S. cerevisiae) |
| chr11_-_90002881 | 0.17 |
ENSMUST00000020864.8
|
Pctp
|
phosphatidylcholine transfer protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of CCCUGAG
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.3 | 0.9 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.3 | 1.2 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.3 | 2.6 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 1.7 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.2 | 0.7 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.2 | 2.3 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.2 | 0.8 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.2 | 0.6 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.2 | 0.6 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.2 | 0.6 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.1 | 0.6 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.4 | GO:0006222 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 0.4 | GO:0002352 | B cell negative selection(GO:0002352) |
| 0.1 | 0.5 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.4 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 0.8 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.1 | 0.4 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.3 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.4 | GO:1903898 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.1 | 0.5 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.5 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 1.3 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.1 | 0.3 | GO:0072573 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.1 | 0.4 | GO:0060031 | mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.1 | 0.3 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.4 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.3 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.3 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) bile acid signaling pathway(GO:0038183) |
| 0.1 | 0.7 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.4 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.6 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 1.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.3 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 0.2 | GO:0009838 | abscission(GO:0009838) |
| 0.1 | 1.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 1.4 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.1 | 0.4 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.5 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.1 | 0.6 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.3 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.2 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.2 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.5 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.2 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) |
| 0.1 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.2 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) |
| 0.1 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 0.2 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.2 | GO:0098917 | diacylglycerol catabolic process(GO:0046340) retrograde trans-synaptic signaling(GO:0098917) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.0 | 0.8 | GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:1990168 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.5 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.2 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.2 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.7 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.5 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.3 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.3 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.3 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0015920 | regulation of phosphatidylcholine catabolic process(GO:0010899) lipopolysaccharide transport(GO:0015920) |
| 0.0 | 0.3 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.1 | GO:0002215 | defense response to nematode(GO:0002215) memory T cell activation(GO:0035709) positive regulation of memory T cell differentiation(GO:0043382) negative regulation of T-helper 1 cell differentiation(GO:0045626) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.3 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.4 | GO:0032274 | gonadotropin secretion(GO:0032274) |
| 0.0 | 0.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.3 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.3 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.3 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.1 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 | 0.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.0 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.2 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) nuclear matrix anchoring at nuclear membrane(GO:0090292) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.3 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.3 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.3 | GO:0034377 | plasma lipoprotein particle assembly(GO:0034377) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.3 | GO:0032695 | positive regulation of macrophage chemotaxis(GO:0010759) negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.0 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.5 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.2 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.1 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.2 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.1 | 1.0 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 0.3 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.1 | 0.3 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 0.7 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 0.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.6 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.3 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 1.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 1.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.5 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 1.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.5 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 1.0 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.4 | GO:0090544 | BAF-type complex(GO:0090544) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.2 | 1.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 0.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 0.8 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 1.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 0.6 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.1 | 1.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.8 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 1.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.4 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.1 | 0.8 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 0.3 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.1 | 0.3 | GO:1902271 | lithocholic acid binding(GO:1902121) D3 vitamins binding(GO:1902271) |
| 0.1 | 0.4 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 1.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.4 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.7 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 0.5 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.3 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.2 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 2.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.3 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.2 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.3 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.0 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.2 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 2.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 2.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.5 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 0.8 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 1.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |