Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
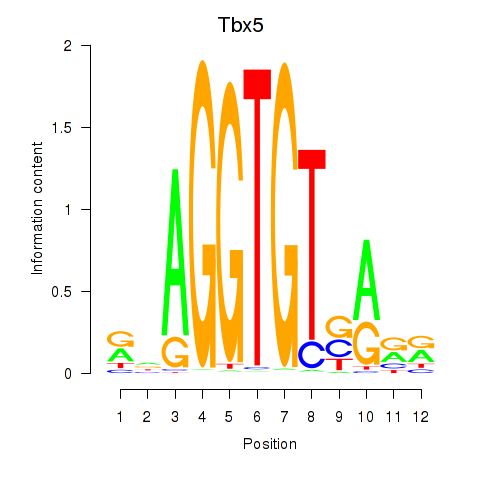
Results for Tbx5
Z-value: 1.37
Transcription factors associated with Tbx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx5
|
ENSMUSG00000018263.8 | T-box 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx5 | mm10_v2_chr5_+_119834663_119834663 | 0.28 | 9.8e-02 | Click! |
Activity profile of Tbx5 motif
Sorted Z-values of Tbx5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_80000292 | 10.98 |
ENSMUST00000088735.3
|
Olfm4
|
olfactomedin 4 |
| chr10_+_75566257 | 10.95 |
ENSMUST00000129232.1
ENSMUST00000143792.1 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr8_+_23035116 | 9.84 |
ENSMUST00000117296.1
ENSMUST00000141784.2 |
Ank1
|
ankyrin 1, erythroid |
| chr4_-_134018829 | 9.18 |
ENSMUST00000051674.2
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr11_-_87875524 | 8.30 |
ENSMUST00000049768.3
|
Epx
|
eosinophil peroxidase |
| chr7_-_142666816 | 8.04 |
ENSMUST00000105935.1
|
Igf2
|
insulin-like growth factor 2 |
| chr15_-_89425856 | 7.81 |
ENSMUST00000109313.2
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr15_-_103255433 | 7.69 |
ENSMUST00000075192.6
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr1_-_132390301 | 6.98 |
ENSMUST00000132435.1
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr3_-_20275659 | 6.92 |
ENSMUST00000011607.5
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr14_-_56085214 | 6.83 |
ENSMUST00000015594.7
|
Mcpt8
|
mast cell protease 8 |
| chr8_-_105933832 | 6.67 |
ENSMUST00000034368.6
|
Ctrl
|
chymotrypsin-like |
| chr6_+_30639218 | 6.43 |
ENSMUST00000031806.9
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr19_+_54045182 | 6.19 |
ENSMUST00000036700.5
|
Adra2a
|
adrenergic receptor, alpha 2a |
| chr3_-_100489324 | 5.77 |
ENSMUST00000061455.8
|
Fam46c
|
family with sequence similarity 46, member C |
| chr14_+_55765956 | 5.75 |
ENSMUST00000057569.3
|
Ltb4r1
|
leukotriene B4 receptor 1 |
| chr4_+_115059507 | 5.62 |
ENSMUST00000162489.1
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr5_-_148399901 | 5.57 |
ENSMUST00000048116.8
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr11_-_69900930 | 5.48 |
ENSMUST00000018714.6
ENSMUST00000128046.1 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr11_-_46312220 | 5.14 |
ENSMUST00000129474.1
ENSMUST00000093166.4 ENSMUST00000165599.2 |
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr15_-_89425795 | 5.13 |
ENSMUST00000168376.1
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr11_-_69900949 | 5.10 |
ENSMUST00000102580.3
|
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr11_-_116076986 | 4.94 |
ENSMUST00000153408.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr2_+_103970115 | 4.89 |
ENSMUST00000111143.1
ENSMUST00000138815.1 |
Lmo2
|
LIM domain only 2 |
| chr1_-_193035651 | 4.86 |
ENSMUST00000016344.7
|
Syt14
|
synaptotagmin XIV |
| chr1_+_135133272 | 4.69 |
ENSMUST00000167080.1
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr14_-_51057242 | 4.55 |
ENSMUST00000089798.2
|
Rnase12
|
ribonuclease, RNase A family, 12 (non-active) |
| chr9_-_95845215 | 4.52 |
ENSMUST00000093800.2
|
Pls1
|
plastin 1 (I-isoform) |
| chr7_+_113513854 | 4.43 |
ENSMUST00000067929.8
ENSMUST00000129087.1 ENSMUST00000164745.1 ENSMUST00000136158.1 |
Far1
|
fatty acyl CoA reductase 1 |
| chr4_+_154869585 | 4.28 |
ENSMUST00000079269.7
ENSMUST00000163732.1 ENSMUST00000080559.6 |
Mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr2_+_103970221 | 4.27 |
ENSMUST00000111140.2
ENSMUST00000111139.2 |
Lmo2
|
LIM domain only 2 |
| chr3_+_108383829 | 4.18 |
ENSMUST00000090561.3
ENSMUST00000102629.1 ENSMUST00000128089.1 |
Psrc1
|
proline/serine-rich coiled-coil 1 |
| chrX_+_100625737 | 4.10 |
ENSMUST00000048962.3
|
Kif4
|
kinesin family member 4 |
| chr11_+_72961163 | 4.08 |
ENSMUST00000108486.1
ENSMUST00000108484.1 ENSMUST00000021142.7 ENSMUST00000108485.2 ENSMUST00000163326.1 |
Atp2a3
|
ATPase, Ca++ transporting, ubiquitous |
| chr7_+_28982832 | 3.92 |
ENSMUST00000085835.6
|
Map4k1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chrX_-_49797700 | 3.89 |
ENSMUST00000033442.7
ENSMUST00000114891.1 |
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr11_+_69095217 | 3.77 |
ENSMUST00000101004.2
|
Per1
|
period circadian clock 1 |
| chr1_+_91366412 | 3.70 |
ENSMUST00000086861.5
|
Fam132b
|
family with sequence similarity 132, member B |
| chr2_-_181691771 | 3.62 |
ENSMUST00000108778.1
ENSMUST00000165416.1 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr2_-_32387760 | 3.61 |
ENSMUST00000050785.8
|
Lcn2
|
lipocalin 2 |
| chr7_+_113513829 | 3.57 |
ENSMUST00000033018.8
|
Far1
|
fatty acyl CoA reductase 1 |
| chr7_+_81858993 | 3.53 |
ENSMUST00000041890.1
|
Tm6sf1
|
transmembrane 6 superfamily member 1 |
| chr7_-_45239108 | 3.52 |
ENSMUST00000033063.6
|
Cd37
|
CD37 antigen |
| chr11_+_53519725 | 3.44 |
ENSMUST00000108987.1
ENSMUST00000121334.1 ENSMUST00000117061.1 |
Sept8
|
septin 8 |
| chr7_-_141117772 | 3.32 |
ENSMUST00000067836.7
|
Ano9
|
anoctamin 9 |
| chr6_-_49214954 | 3.31 |
ENSMUST00000031838.7
|
Igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr8_-_121907678 | 3.31 |
ENSMUST00000045557.9
|
Slc7a5
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 |
| chr5_-_137741601 | 3.22 |
ENSMUST00000119498.1
ENSMUST00000061789.7 |
Nyap1
|
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
| chr8_-_105637403 | 3.20 |
ENSMUST00000182046.1
|
Gm5914
|
predicted gene 5914 |
| chrX_-_36989656 | 3.15 |
ENSMUST00000060474.7
ENSMUST00000053456.4 ENSMUST00000115239.3 |
Sept6
|
septin 6 |
| chr7_-_45239041 | 3.12 |
ENSMUST00000131290.1
|
Cd37
|
CD37 antigen |
| chr15_+_79347534 | 2.96 |
ENSMUST00000096350.3
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr9_+_108991902 | 2.95 |
ENSMUST00000147989.1
ENSMUST00000051873.8 |
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr18_-_54990124 | 2.93 |
ENSMUST00000064763.5
|
Zfp608
|
zinc finger protein 608 |
| chr3_+_123446913 | 2.86 |
ENSMUST00000029603.8
|
Prss12
|
protease, serine, 12 neurotrypsin (motopsin) |
| chr7_-_45238794 | 2.84 |
ENSMUST00000098461.1
ENSMUST00000107797.1 |
Cd37
|
CD37 antigen |
| chr14_+_65266701 | 2.81 |
ENSMUST00000169656.1
|
Fbxo16
|
F-box protein 16 |
| chr8_-_85119637 | 2.80 |
ENSMUST00000098550.3
|
Zfp791
|
zinc finger protein 791 |
| chrX_+_134295225 | 2.80 |
ENSMUST00000037687.7
|
Tmem35
|
transmembrane protein 35 |
| chr7_-_104950441 | 2.70 |
ENSMUST00000179862.1
|
Gm5900
|
predicted pseudogene 5900 |
| chr17_-_25952565 | 2.63 |
ENSMUST00000162431.1
|
A930017K11Rik
|
RIKEN cDNA A930017K11 gene |
| chr3_+_103102604 | 2.62 |
ENSMUST00000173206.1
|
Dennd2c
|
DENN/MADD domain containing 2C |
| chr9_+_86743641 | 2.61 |
ENSMUST00000179574.1
|
Prss35
|
protease, serine, 35 |
| chr5_-_137741102 | 2.58 |
ENSMUST00000149512.1
|
Nyap1
|
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
| chr9_+_75232013 | 2.58 |
ENSMUST00000036555.6
|
Myo5c
|
myosin VC |
| chr12_-_113422730 | 2.57 |
ENSMUST00000177715.1
ENSMUST00000103426.1 |
Ighm
|
immunoglobulin heavy constant mu |
| chr19_-_46327121 | 2.54 |
ENSMUST00000041391.4
ENSMUST00000096029.5 |
Psd
|
pleckstrin and Sec7 domain containing |
| chrX_+_159840463 | 2.52 |
ENSMUST00000112451.1
ENSMUST00000112453.2 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr5_+_143622466 | 2.51 |
ENSMUST00000177196.1
|
Cyth3
|
cytohesin 3 |
| chr3_+_130180882 | 2.46 |
ENSMUST00000106353.1
ENSMUST00000080335.4 |
Col25a1
|
collagen, type XXV, alpha 1 |
| chr15_-_83033294 | 2.35 |
ENSMUST00000100377.4
|
Nfam1
|
Nfat activating molecule with ITAM motif 1 |
| chr4_+_103619580 | 2.34 |
ENSMUST00000106827.1
|
Dab1
|
disabled 1 |
| chr17_+_47505211 | 2.32 |
ENSMUST00000182935.1
ENSMUST00000182506.1 |
Ccnd3
|
cyclin D3 |
| chr9_+_73044833 | 2.30 |
ENSMUST00000184146.1
ENSMUST00000034722.4 |
Rab27a
|
RAB27A, member RAS oncogene family |
| chr8_+_120537423 | 2.30 |
ENSMUST00000118136.1
|
Gse1
|
genetic suppressor element 1 |
| chr11_-_8973266 | 2.26 |
ENSMUST00000154153.1
|
Pkd1l1
|
polycystic kidney disease 1 like 1 |
| chr7_-_141443989 | 2.25 |
ENSMUST00000026580.5
|
Lrdd
|
leucine-rich and death domain containing |
| chr9_+_86743616 | 2.25 |
ENSMUST00000036426.6
|
Prss35
|
protease, serine, 35 |
| chr5_-_24329556 | 2.22 |
ENSMUST00000115098.2
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr19_+_38931008 | 2.19 |
ENSMUST00000145051.1
|
Hells
|
helicase, lymphoid specific |
| chr17_+_47505043 | 2.16 |
ENSMUST00000182129.1
ENSMUST00000171031.1 |
Ccnd3
|
cyclin D3 |
| chr4_-_43045686 | 2.15 |
ENSMUST00000107956.1
ENSMUST00000107957.1 |
Fam214b
|
family with sequence similarity 214, member B |
| chr11_+_96929367 | 2.12 |
ENSMUST00000062172.5
|
Prr15l
|
proline rich 15-like |
| chrX_-_51205990 | 2.12 |
ENSMUST00000114876.2
|
Mbnl3
|
muscleblind-like 3 (Drosophila) |
| chr11_-_106301801 | 2.11 |
ENSMUST00000103071.3
|
Gh
|
growth hormone |
| chr10_+_60346851 | 2.09 |
ENSMUST00000020301.7
ENSMUST00000105460.1 ENSMUST00000170507.1 |
4632428N05Rik
|
RIKEN cDNA 4632428N05 gene |
| chr19_+_38097065 | 2.08 |
ENSMUST00000067098.6
|
Ffar4
|
free fatty acid receptor 4 |
| chr17_+_47505149 | 2.07 |
ENSMUST00000183177.1
ENSMUST00000182848.1 |
Ccnd3
|
cyclin D3 |
| chr17_+_47505117 | 2.06 |
ENSMUST00000183044.1
ENSMUST00000037333.10 |
Ccnd3
|
cyclin D3 |
| chr10_-_91082653 | 2.06 |
ENSMUST00000159110.1
|
Apaf1
|
apoptotic peptidase activating factor 1 |
| chr5_+_143622440 | 2.06 |
ENSMUST00000116456.3
|
Cyth3
|
cytohesin 3 |
| chr4_+_58943575 | 2.00 |
ENSMUST00000107554.1
|
Zkscan16
|
zinc finger with KRAB and SCAN domains 16 |
| chr3_-_126998408 | 2.00 |
ENSMUST00000182764.1
ENSMUST00000044443.8 |
Ank2
|
ankyrin 2, brain |
| chr7_-_4546567 | 2.00 |
ENSMUST00000065957.5
|
Syt5
|
synaptotagmin V |
| chr8_-_68121527 | 1.99 |
ENSMUST00000178529.1
|
Gm21807
|
predicted gene, 21807 |
| chr7_+_141131268 | 1.99 |
ENSMUST00000026568.8
|
Ptdss2
|
phosphatidylserine synthase 2 |
| chr3_-_65529355 | 1.97 |
ENSMUST00000099076.3
|
4931440P22Rik
|
RIKEN cDNA 4931440P22 gene |
| chr15_-_75888754 | 1.97 |
ENSMUST00000184858.1
|
Mroh6
|
maestro heat-like repeat family member 6 |
| chr7_+_43437073 | 1.96 |
ENSMUST00000070518.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr19_-_10304867 | 1.93 |
ENSMUST00000039327.4
|
Dagla
|
diacylglycerol lipase, alpha |
| chr2_-_54085542 | 1.93 |
ENSMUST00000100089.2
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr17_+_47726834 | 1.92 |
ENSMUST00000024782.5
ENSMUST00000144955.1 |
Pgc
|
progastricsin (pepsinogen C) |
| chrX_-_51205773 | 1.90 |
ENSMUST00000114875.1
|
Mbnl3
|
muscleblind-like 3 (Drosophila) |
| chr15_-_99457712 | 1.90 |
ENSMUST00000161948.1
|
Nckap5l
|
NCK-associated protein 5-like |
| chr1_+_66364623 | 1.88 |
ENSMUST00000077355.5
ENSMUST00000114012.1 |
Map2
|
microtubule-associated protein 2 |
| chr11_-_69900886 | 1.80 |
ENSMUST00000108621.2
ENSMUST00000100969.2 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr10_-_91082704 | 1.80 |
ENSMUST00000162618.1
ENSMUST00000020157.6 ENSMUST00000160788.1 |
Apaf1
|
apoptotic peptidase activating factor 1 |
| chr4_-_117883428 | 1.80 |
ENSMUST00000030266.5
|
B4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr16_+_17276291 | 1.79 |
ENSMUST00000164950.1
ENSMUST00000159242.1 |
Tmem191c
|
transmembrane protein 191C |
| chr8_+_83389846 | 1.76 |
ENSMUST00000002259.6
|
Clgn
|
calmegin |
| chr6_-_52158292 | 1.76 |
ENSMUST00000000964.5
ENSMUST00000120363.1 |
Hoxa1
|
homeobox A1 |
| chrX_+_100729917 | 1.74 |
ENSMUST00000019503.7
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr7_-_102100227 | 1.74 |
ENSMUST00000106937.1
|
Art5
|
ADP-ribosyltransferase 5 |
| chr9_-_35558522 | 1.74 |
ENSMUST00000034612.5
|
Ddx25
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 25 |
| chr1_+_87620306 | 1.72 |
ENSMUST00000169754.1
|
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chrX_-_7740206 | 1.72 |
ENSMUST00000128289.1
|
Ccdc120
|
coiled-coil domain containing 120 |
| chr8_+_83389878 | 1.69 |
ENSMUST00000109831.2
|
Clgn
|
calmegin |
| chr15_-_27681498 | 1.69 |
ENSMUST00000100739.3
|
Fam105a
|
family with sequence similarity 105, member A |
| chr16_+_17276337 | 1.68 |
ENSMUST00000159065.1
ENSMUST00000159494.1 ENSMUST00000159811.1 |
Tmem191c
|
transmembrane protein 191C |
| chr11_+_69965396 | 1.68 |
ENSMUST00000018713.6
|
Cldn7
|
claudin 7 |
| chr4_+_132564051 | 1.68 |
ENSMUST00000070690.7
|
Ptafr
|
platelet-activating factor receptor |
| chr19_+_38930909 | 1.67 |
ENSMUST00000025965.5
|
Hells
|
helicase, lymphoid specific |
| chr8_-_105637350 | 1.67 |
ENSMUST00000182863.1
|
Gm5914
|
predicted gene 5914 |
| chr3_-_127225917 | 1.64 |
ENSMUST00000182064.1
ENSMUST00000182662.1 |
Ank2
|
ankyrin 2, brain |
| chr9_+_107580746 | 1.63 |
ENSMUST00000148440.1
|
Hyal3
|
hyaluronoglucosaminidase 3 |
| chr11_+_98383811 | 1.63 |
ENSMUST00000008021.2
|
Tcap
|
titin-cap |
| chr2_-_44927161 | 1.59 |
ENSMUST00000130991.1
|
Gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr3_-_127225847 | 1.58 |
ENSMUST00000182726.1
ENSMUST00000182779.1 |
Ank2
|
ankyrin 2, brain |
| chr2_+_121358591 | 1.58 |
ENSMUST00000000317.6
ENSMUST00000129130.1 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chrX_+_135885851 | 1.56 |
ENSMUST00000180025.1
ENSMUST00000068755.7 ENSMUST00000148374.1 |
Bhlhb9
|
basic helix-loop-helix domain containing, class B9 |
| chr6_+_48718602 | 1.52 |
ENSMUST00000127537.1
ENSMUST00000155017.2 |
RP24-315D19.2
|
GTPase IMAP family member 1 |
| chr5_-_22344690 | 1.52 |
ENSMUST00000062372.7
ENSMUST00000161356.1 |
Reln
|
reelin |
| chr15_-_99457742 | 1.52 |
ENSMUST00000023747.7
|
Nckap5l
|
NCK-associated protein 5-like |
| chr11_+_78188422 | 1.51 |
ENSMUST00000002128.7
ENSMUST00000150941.1 |
Rab34
|
RAB34, member of RAS oncogene family |
| chr2_-_44927206 | 1.51 |
ENSMUST00000100127.2
|
Gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr7_-_45824750 | 1.49 |
ENSMUST00000071937.5
|
Kcnj14
|
potassium inwardly-rectifying channel, subfamily J, member 14 |
| chrX_-_147554050 | 1.48 |
ENSMUST00000112819.2
ENSMUST00000136789.1 |
Lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr6_+_35252692 | 1.47 |
ENSMUST00000130875.1
|
1810058I24Rik
|
RIKEN cDNA 1810058I24 gene |
| chr11_+_96929260 | 1.46 |
ENSMUST00000054311.5
ENSMUST00000107636.3 |
Prr15l
|
proline rich 15-like |
| chr1_+_135836380 | 1.45 |
ENSMUST00000178204.1
|
Tnnt2
|
troponin T2, cardiac |
| chr2_+_121357714 | 1.44 |
ENSMUST00000125812.1
ENSMUST00000078222.2 ENSMUST00000125221.1 ENSMUST00000150271.1 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr11_-_6606053 | 1.44 |
ENSMUST00000045713.3
|
Nacad
|
NAC alpha domain containing |
| chr6_+_35252654 | 1.43 |
ENSMUST00000152147.1
|
1810058I24Rik
|
RIKEN cDNA 1810058I24 gene |
| chr7_+_123214776 | 1.42 |
ENSMUST00000131933.1
|
Slc5a11
|
solute carrier family 5 (sodium/glucose cotransporter), member 11 |
| chr17_+_48264270 | 1.42 |
ENSMUST00000059873.7
ENSMUST00000154335.1 ENSMUST00000136272.1 ENSMUST00000125426.1 ENSMUST00000153420.1 |
Treml4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr11_+_54304180 | 1.41 |
ENSMUST00000108904.3
ENSMUST00000108905.3 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr6_-_56901870 | 1.39 |
ENSMUST00000101367.2
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr6_+_35252724 | 1.37 |
ENSMUST00000136110.1
|
1810058I24Rik
|
RIKEN cDNA 1810058I24 gene |
| chr11_+_54304005 | 1.37 |
ENSMUST00000000145.5
ENSMUST00000138515.1 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr6_+_17307040 | 1.34 |
ENSMUST00000123439.1
|
Cav1
|
caveolin 1, caveolae protein |
| chr6_+_115134899 | 1.34 |
ENSMUST00000009538.5
ENSMUST00000169345.1 |
Syn2
|
synapsin II |
| chr6_-_28831747 | 1.33 |
ENSMUST00000062304.5
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr4_+_119814495 | 1.32 |
ENSMUST00000106307.2
|
Hivep3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr6_+_41684414 | 1.31 |
ENSMUST00000031900.5
|
1700034O15Rik
|
RIKEN cDNA 1700034O15 gene |
| chr17_+_5975740 | 1.31 |
ENSMUST00000115790.1
|
Synj2
|
synaptojanin 2 |
| chr17_+_47505073 | 1.30 |
ENSMUST00000183210.1
|
Ccnd3
|
cyclin D3 |
| chr8_-_80739497 | 1.30 |
ENSMUST00000043359.8
|
Smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr11_+_96323253 | 1.30 |
ENSMUST00000093944.3
|
Hoxb3
|
homeobox B3 |
| chrX_-_136085206 | 1.29 |
ENSMUST00000138878.1
ENSMUST00000080929.6 |
Nxf3
|
nuclear RNA export factor 3 |
| chrX_+_100730178 | 1.29 |
ENSMUST00000113744.1
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr7_-_115824699 | 1.26 |
ENSMUST00000169129.1
|
Sox6
|
SRY-box containing gene 6 |
| chr12_+_51348370 | 1.26 |
ENSMUST00000121521.1
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chr11_-_70220776 | 1.24 |
ENSMUST00000141290.1
|
Slc16a13
|
solute carrier family 16 (monocarboxylic acid transporters), member 13 |
| chr9_-_59146210 | 1.23 |
ENSMUST00000085631.2
|
Gm7589
|
predicted gene 7589 |
| chr17_+_21383725 | 1.23 |
ENSMUST00000056107.4
ENSMUST00000162659.1 |
Zfp677
|
zinc finger protein 677 |
| chr7_+_25267669 | 1.23 |
ENSMUST00000169266.1
|
Cic
|
capicua homolog (Drosophila) |
| chr15_+_103453782 | 1.21 |
ENSMUST00000047405.7
|
Nckap1l
|
NCK associated protein 1 like |
| chr12_-_27342696 | 1.19 |
ENSMUST00000079063.5
|
Sox11
|
SRY-box containing gene 11 |
| chr2_+_24186469 | 1.19 |
ENSMUST00000057567.2
|
Il1f9
|
interleukin 1 family, member 9 |
| chr7_-_35396708 | 1.19 |
ENSMUST00000154597.1
ENSMUST00000032704.5 |
C230052I12Rik
|
RIKEN cDNA C230052I12 gene |
| chr7_-_140082246 | 1.16 |
ENSMUST00000166758.2
|
Caly
|
calcyon neuron-specific vesicular protein |
| chr6_+_4504814 | 1.16 |
ENSMUST00000141483.1
|
Col1a2
|
collagen, type I, alpha 2 |
| chr17_+_32036098 | 1.16 |
ENSMUST00000081339.6
|
Rrp1b
|
ribosomal RNA processing 1 homolog B (S. cerevisiae) |
| chr11_-_120598346 | 1.15 |
ENSMUST00000026125.2
|
Alyref
|
Aly/REF export factor |
| chr15_-_63997969 | 1.15 |
ENSMUST00000164532.1
|
Fam49b
|
family with sequence similarity 49, member B |
| chr1_-_178337774 | 1.15 |
ENSMUST00000037748.7
|
Hnrnpu
|
heterogeneous nuclear ribonucleoprotein U |
| chr11_+_20647149 | 1.14 |
ENSMUST00000109585.1
|
Sertad2
|
SERTA domain containing 2 |
| chr5_-_143292356 | 1.14 |
ENSMUST00000180336.1
|
Zfp853
|
zinc finger protein 853 |
| chr12_+_51348019 | 1.14 |
ENSMUST00000054308.6
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chrX_+_42151002 | 1.12 |
ENSMUST00000123245.1
|
Stag2
|
stromal antigen 2 |
| chr6_+_54681687 | 1.12 |
ENSMUST00000046276.6
|
2410066E13Rik
|
RIKEN cDNA 2410066E13 gene |
| chr11_-_70220794 | 1.12 |
ENSMUST00000159867.1
|
Slc16a13
|
solute carrier family 16 (monocarboxylic acid transporters), member 13 |
| chr1_-_167285110 | 1.12 |
ENSMUST00000027839.8
|
Uck2
|
uridine-cytidine kinase 2 |
| chr8_+_125669818 | 1.09 |
ENSMUST00000053078.3
|
Map10
|
microtubule-associated protein 10 |
| chr7_-_44748617 | 1.09 |
ENSMUST00000060270.6
|
Zfp473
|
zinc finger protein 473 |
| chr9_-_96437434 | 1.09 |
ENSMUST00000070500.2
|
BC043934
|
cDNA sequence BC043934 |
| chr13_+_118714678 | 1.06 |
ENSMUST00000022246.8
|
Fgf10
|
fibroblast growth factor 10 |
| chr1_+_63273261 | 1.06 |
ENSMUST00000114132.1
ENSMUST00000126932.1 |
Zdbf2
|
zinc finger, DBF-type containing 2 |
| chr3_+_126597299 | 1.06 |
ENSMUST00000106400.2
ENSMUST00000106401.1 |
Camk2d
|
calcium/calmodulin-dependent protein kinase II, delta |
| chr10_-_14544972 | 1.03 |
ENSMUST00000041168.4
|
Gpr126
|
G protein-coupled receptor 126 |
| chr11_-_70220969 | 1.03 |
ENSMUST00000060010.2
|
Slc16a13
|
solute carrier family 16 (monocarboxylic acid transporters), member 13 |
| chr3_-_95306794 | 1.03 |
ENSMUST00000107183.1
ENSMUST00000164406.1 ENSMUST00000123365.1 |
Anxa9
|
annexin A9 |
| chr5_+_113490447 | 1.03 |
ENSMUST00000094452.3
|
Wscd2
|
WSC domain containing 2 |
| chr18_+_82914632 | 0.99 |
ENSMUST00000071233.6
|
Zfp516
|
zinc finger protein 516 |
| chr6_-_28830345 | 0.99 |
ENSMUST00000171353.1
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr1_+_93373874 | 0.98 |
ENSMUST00000058682.4
|
Ano7
|
anoctamin 7 |
| chr5_+_17574726 | 0.98 |
ENSMUST00000169603.1
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr18_+_36793918 | 0.98 |
ENSMUST00000001419.8
|
Zmat2
|
zinc finger, matrin type 2 |
| chr19_-_6925367 | 0.96 |
ENSMUST00000057716.4
|
Tex40
|
testis expressed 40 |
| chr7_-_113369326 | 0.94 |
ENSMUST00000047091.7
ENSMUST00000119278.1 |
Btbd10
|
BTB (POZ) domain containing 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.3 | GO:0002215 | defense response to nematode(GO:0002215) |
| 1.9 | 5.6 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 1.4 | 10.9 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 1.3 | 8.0 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 1.1 | 4.5 | GO:1902896 | terminal web assembly(GO:1902896) |
| 1.1 | 10.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 1.1 | 3.3 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 1.0 | 6.2 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 1.0 | 8.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 1.0 | 3.9 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.9 | 9.5 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.9 | 2.6 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.8 | 3.8 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.8 | 2.3 | GO:0003127 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.7 | 8.4 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.6 | 9.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.6 | 1.8 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.6 | 5.2 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.6 | 1.1 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.6 | 1.1 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.6 | 1.7 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.6 | 2.2 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.6 | 3.9 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.5 | 4.9 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.5 | 5.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.5 | 3.6 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.5 | 4.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.5 | 2.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.5 | 2.5 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.5 | 1.9 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) retrograde trans-synaptic signaling(GO:0098917) |
| 0.5 | 3.9 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.5 | 1.4 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.5 | 2.8 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.5 | 2.3 | GO:0097278 | complement-dependent cytotoxicity(GO:0097278) |
| 0.5 | 1.8 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.4 | 9.9 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.4 | 1.7 | GO:0045659 | negative regulation of neutrophil differentiation(GO:0045659) |
| 0.4 | 2.1 | GO:1905169 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.4 | 1.9 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.4 | 1.1 | GO:0006222 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.4 | 2.2 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.4 | 1.1 | GO:0070350 | bronchiole development(GO:0060435) secretion by lung epithelial cell involved in lung growth(GO:0061033) white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.4 | 2.5 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.3 | 5.6 | GO:0015809 | arginine transport(GO:0015809) |
| 0.3 | 2.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 0.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.3 | 3.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.3 | 1.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 | 2.3 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.3 | 1.6 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.3 | 1.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.3 | 11.1 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.3 | 0.5 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.2 | 1.0 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.2 | 1.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.2 | 0.7 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.2 | 1.4 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.2 | 0.7 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 11.0 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 6.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 0.6 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.2 | 2.3 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.2 | 1.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.2 | 0.7 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 0.5 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.2 | 0.2 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.2 | 0.7 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.2 | 1.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.2 | 13.2 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.2 | 0.5 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.2 | 0.5 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.2 | 0.9 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.2 | 0.3 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.1 | 2.1 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.1 | 2.6 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.1 | 0.4 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.1 | 0.4 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 1.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.9 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.1 | 0.3 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 1.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.6 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.4 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.1 | 1.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 4.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.7 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 3.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 6.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 1.4 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.3 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 1.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.4 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.3 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 0.1 | 0.5 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 0.5 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 0.6 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.1 | 5.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 4.7 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 0.7 | GO:0071816 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 1.7 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.3 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 1.8 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.6 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 1.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 3.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.4 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 1.2 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 | 5.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 0.2 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.1 | 1.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.3 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 2.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 1.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.8 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.6 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.6 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 4.5 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 1.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.6 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 2.1 | GO:0050710 | negative regulation of cytokine secretion(GO:0050710) |
| 0.0 | 5.3 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 4.2 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.8 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.9 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.8 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.0 | 4.9 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 4.1 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.3 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.6 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.4 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 1.0 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.9 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.5 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 1.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.3 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 1.2 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.4 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.0 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 1.0 | 3.1 | GO:0005940 | septin ring(GO:0005940) |
| 1.0 | 3.9 | GO:0043293 | apoptosome(GO:0043293) |
| 0.9 | 12.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.7 | 11.0 | GO:0042581 | specific granule(GO:0042581) |
| 0.7 | 7.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.6 | 4.5 | GO:1990357 | terminal web(GO:1990357) |
| 0.6 | 2.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.4 | 1.3 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.4 | 9.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.4 | 2.0 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.4 | 1.2 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.4 | 2.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.3 | 8.0 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.2 | 1.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 3.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 1.9 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 0.6 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.2 | 1.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 2.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 9.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 5.6 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.8 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 0.6 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.1 | 1.5 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.7 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 1.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 7.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.3 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 3.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.7 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 1.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 9.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.9 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.8 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 1.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 4.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 1.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 11.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.6 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 6.7 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 2.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 1.0 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 6.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 2.3 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 1.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.7 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 4.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 7.4 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 2.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 5.8 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 3.0 | GO:0044455 | mitochondrial membrane part(GO:0044455) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 20.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.4 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 1.3 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 12.9 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 1.6 | 8.0 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 1.5 | 6.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 1.4 | 5.7 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 1.0 | 9.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.9 | 10.9 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.8 | 9.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.7 | 5.6 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.6 | 2.4 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.6 | 1.8 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.5 | 1.6 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.5 | 3.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.5 | 2.5 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.4 | 3.9 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.4 | 13.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 1.1 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.4 | 2.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.3 | 3.0 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.3 | 1.7 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.3 | 2.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.3 | 1.6 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.3 | 16.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 1.7 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.3 | 1.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.3 | 2.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.3 | 1.5 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.2 | 2.0 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.2 | 0.7 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.2 | 8.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 3.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.2 | 2.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 1.9 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 4.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 2.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 1.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 2.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.2 | 1.4 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 9.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 0.9 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.2 | 6.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 8.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 0.7 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.2 | 7.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 0.5 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.2 | 3.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 1.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 2.8 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.8 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 3.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.9 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 4.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 8.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 1.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.8 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.5 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 6.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 2.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 2.1 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 2.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.6 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 1.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 1.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 4.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.4 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 3.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.5 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 3.0 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 1.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 2.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 2.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.3 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 6.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 6.4 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 4.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 3.9 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 3.4 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 3.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.2 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 1.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.7 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 1.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 4.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 3.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 3.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 8.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 2.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 6.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 8.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 4.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.5 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 2.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 3.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.4 | 8.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 10.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 8.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 9.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.2 | 9.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 6.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.0 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 0.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 2.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 3.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 2.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 3.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 2.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 2.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 2.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 3.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 2.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 3.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 7.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.8 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 2.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.9 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |