Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
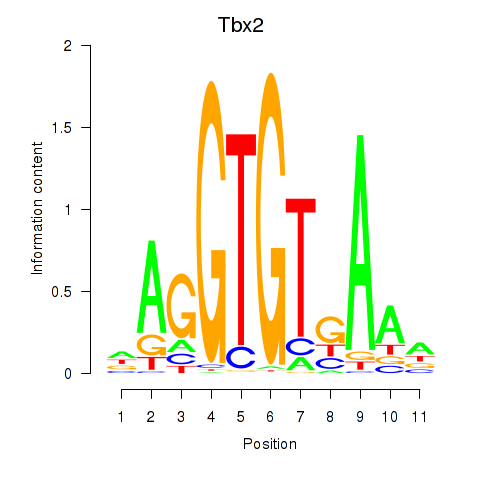
Results for Tbx2
Z-value: 1.35
Transcription factors associated with Tbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx2
|
ENSMUSG00000000093.6 | T-box 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx2 | mm10_v2_chr11_+_85832551_85832551 | -0.63 | 3.9e-05 | Click! |
Activity profile of Tbx2 motif
Sorted Z-values of Tbx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_130826676 | 18.75 |
ENSMUST00000027675.7
|
Pigr
|
polymeric immunoglobulin receptor |
| chr1_+_130826762 | 17.86 |
ENSMUST00000133792.1
|
Pigr
|
polymeric immunoglobulin receptor |
| chr6_-_141856171 | 13.53 |
ENSMUST00000165990.1
ENSMUST00000163678.1 |
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr6_+_122006798 | 12.95 |
ENSMUST00000081777.6
|
Mug2
|
murinoglobulin 2 |
| chr19_+_37697792 | 10.45 |
ENSMUST00000025946.5
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr15_+_6445320 | 9.32 |
ENSMUST00000022749.9
|
C9
|
complement component 9 |
| chr10_+_79890853 | 7.88 |
ENSMUST00000061653.7
|
Cfd
|
complement factor D (adipsin) |
| chr16_+_23107413 | 5.50 |
ENSMUST00000023599.6
ENSMUST00000168891.1 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chrX_-_139782353 | 5.47 |
ENSMUST00000101217.3
|
Ripply1
|
ripply1 homolog (zebrafish) |
| chr9_-_43239816 | 5.10 |
ENSMUST00000034512.5
|
Oaf
|
OAF homolog (Drosophila) |
| chr5_-_151190154 | 5.02 |
ENSMUST00000062015.8
ENSMUST00000110483.2 |
Stard13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr1_-_105356658 | 4.98 |
ENSMUST00000058688.5
ENSMUST00000172299.1 |
Rnf152
|
ring finger protein 152 |
| chr1_-_51915968 | 4.85 |
ENSMUST00000046390.7
|
Myo1b
|
myosin IB |
| chr10_-_24101951 | 4.63 |
ENSMUST00000170267.1
|
Taar8c
|
trace amine-associated receptor 8C |
| chr14_-_30943275 | 4.34 |
ENSMUST00000006704.8
ENSMUST00000163118.1 |
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr2_-_148045891 | 4.24 |
ENSMUST00000109964.1
|
Foxa2
|
forkhead box A2 |
| chr19_-_42202150 | 4.13 |
ENSMUST00000018966.7
|
Sfrp5
|
secreted frizzled-related sequence protein 5 |
| chr3_-_107943705 | 3.98 |
ENSMUST00000106680.1
ENSMUST00000106684.1 ENSMUST00000106685.2 |
Gstm6
|
glutathione S-transferase, mu 6 |
| chr6_+_116264186 | 3.86 |
ENSMUST00000036503.7
ENSMUST00000112900.3 |
Zfand4
|
zinc finger, AN1-type domain 4 |
| chr2_+_160880642 | 3.74 |
ENSMUST00000109456.2
|
Lpin3
|
lipin 3 |
| chr1_+_88138364 | 3.71 |
ENSMUST00000014263.4
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr4_-_103214791 | 3.69 |
ENSMUST00000094947.2
ENSMUST00000036195.6 |
Slc35d1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
| chr10_-_120899067 | 3.58 |
ENSMUST00000092143.5
|
Msrb3
|
methionine sulfoxide reductase B3 |
| chr2_-_175703646 | 3.39 |
ENSMUST00000109027.2
ENSMUST00000179061.1 ENSMUST00000131041.1 |
Gm4245
|
predicted gene 4245 |
| chr7_+_79810727 | 3.31 |
ENSMUST00000107394.1
|
Mesp2
|
mesoderm posterior 2 |
| chr2_+_175372436 | 3.25 |
ENSMUST00000131676.1
ENSMUST00000109048.2 ENSMUST00000109047.2 |
Gm4723
|
predicted gene 4723 |
| chrX_+_101383726 | 3.20 |
ENSMUST00000119190.1
|
Gjb1
|
gap junction protein, beta 1 |
| chr1_+_131970589 | 3.17 |
ENSMUST00000027695.6
|
Slc45a3
|
solute carrier family 45, member 3 |
| chr7_+_45259031 | 3.15 |
ENSMUST00000179310.1
|
Slc6a16
|
solute carrier family 6, member 16 |
| chr5_-_5514730 | 3.06 |
ENSMUST00000115445.1
ENSMUST00000179804.1 ENSMUST00000125110.1 ENSMUST00000115446.1 |
Cldn12
|
claudin 12 |
| chr11_+_103101682 | 3.05 |
ENSMUST00000107040.3
ENSMUST00000140372.1 ENSMUST00000024492.8 ENSMUST00000134884.1 |
Acbd4
|
acyl-Coenzyme A binding domain containing 4 |
| chr17_+_3397189 | 3.02 |
ENSMUST00000072156.6
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr9_-_45204083 | 3.02 |
ENSMUST00000034599.8
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr5_-_108132513 | 2.96 |
ENSMUST00000119784.1
ENSMUST00000117759.1 |
Tmed5
|
transmembrane emp24 protein transport domain containing 5 |
| chr1_-_52500679 | 2.92 |
ENSMUST00000069792.7
|
Nab1
|
Ngfi-A binding protein 1 |
| chr5_-_5514873 | 2.88 |
ENSMUST00000060947.7
|
Cldn12
|
claudin 12 |
| chr19_-_28963863 | 2.85 |
ENSMUST00000161813.1
|
4430402I18Rik
|
RIKEN cDNA 4430402I18 gene |
| chr3_-_115715031 | 2.80 |
ENSMUST00000055676.2
|
S1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr15_-_81360739 | 2.72 |
ENSMUST00000023040.7
|
Slc25a17
|
solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17 |
| chr4_-_49549523 | 2.61 |
ENSMUST00000029987.9
|
Aldob
|
aldolase B, fructose-bisphosphate |
| chr1_-_138847579 | 2.60 |
ENSMUST00000093486.3
ENSMUST00000046870.6 |
Lhx9
|
LIM homeobox protein 9 |
| chrX_+_42502533 | 2.38 |
ENSMUST00000005839.4
|
Sh2d1a
|
SH2 domain protein 1A |
| chr2_+_175469985 | 2.32 |
ENSMUST00000109042.3
ENSMUST00000109002.2 ENSMUST00000109043.2 ENSMUST00000143490.1 |
Gm8923
|
predicted gene 8923 |
| chr4_+_41465134 | 2.28 |
ENSMUST00000030154.6
|
Nudt2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 2 |
| chr4_+_53440516 | 2.16 |
ENSMUST00000107651.2
ENSMUST00000107647.1 |
Slc44a1
|
solute carrier family 44, member 1 |
| chr5_+_120513102 | 2.14 |
ENSMUST00000111889.1
|
Slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr6_-_94700137 | 2.13 |
ENSMUST00000101126.2
ENSMUST00000032105.4 |
Lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr13_-_113180897 | 2.11 |
ENSMUST00000038212.7
|
Gzmk
|
granzyme K |
| chr14_+_51893610 | 2.10 |
ENSMUST00000047726.5
ENSMUST00000161888.1 |
Slc39a2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr1_+_164115264 | 2.10 |
ENSMUST00000162746.1
|
Selp
|
selectin, platelet |
| chr14_-_55643251 | 2.04 |
ENSMUST00000120041.1
ENSMUST00000121937.1 ENSMUST00000133707.1 ENSMUST00000002391.8 ENSMUST00000121791.1 |
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr7_+_105404568 | 2.04 |
ENSMUST00000033187.4
|
Cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr10_+_106470281 | 2.04 |
ENSMUST00000029404.9
ENSMUST00000169303.1 |
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr7_+_27607997 | 2.03 |
ENSMUST00000142365.1
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr6_+_50110837 | 2.03 |
ENSMUST00000167628.1
|
Mpp6
|
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
| chr18_+_84088077 | 2.02 |
ENSMUST00000060223.2
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr3_+_60031754 | 2.01 |
ENSMUST00000029325.3
|
Aadac
|
arylacetamide deacetylase (esterase) |
| chr3_-_84220853 | 1.97 |
ENSMUST00000154152.1
ENSMUST00000107693.2 ENSMUST00000107695.2 |
Trim2
|
tripartite motif-containing 2 |
| chr12_+_84642896 | 1.95 |
ENSMUST00000095551.4
|
Vrtn
|
vertebrae development associated |
| chr13_+_112464070 | 1.91 |
ENSMUST00000183663.1
ENSMUST00000184311.1 ENSMUST00000183886.1 |
Il6st
|
interleukin 6 signal transducer |
| chr2_-_103797617 | 1.91 |
ENSMUST00000028607.6
|
Caprin1
|
cell cycle associated protein 1 |
| chr7_-_28302238 | 1.90 |
ENSMUST00000108315.3
|
Dll3
|
delta-like 3 (Drosophila) |
| chr1_+_131599239 | 1.89 |
ENSMUST00000027690.6
|
Avpr1b
|
arginine vasopressin receptor 1B |
| chr5_-_108132541 | 1.87 |
ENSMUST00000119437.1
ENSMUST00000118036.1 |
Tmed5
|
transmembrane emp24 protein transport domain containing 5 |
| chr13_+_80886095 | 1.86 |
ENSMUST00000161441.1
|
Arrdc3
|
arrestin domain containing 3 |
| chr1_-_157108632 | 1.86 |
ENSMUST00000118207.1
ENSMUST00000027884.6 ENSMUST00000121911.1 |
Tex35
|
testis expressed 35 |
| chr10_+_97693053 | 1.85 |
ENSMUST00000060703.4
|
Ccer1
|
coiled coil glutamate rich protein 1 |
| chr14_-_11162008 | 1.82 |
ENSMUST00000162278.1
ENSMUST00000160340.1 ENSMUST00000160956.1 |
Fhit
|
fragile histidine triad gene |
| chr9_-_48495321 | 1.81 |
ENSMUST00000170000.2
|
Rbm7
|
RNA binding motif protein 7 |
| chr4_+_125490688 | 1.80 |
ENSMUST00000030676.7
|
Grik3
|
glutamate receptor, ionotropic, kainate 3 |
| chr11_-_84068766 | 1.76 |
ENSMUST00000018792.5
|
Dusp14
|
dual specificity phosphatase 14 |
| chr13_+_54503779 | 1.73 |
ENSMUST00000121401.1
ENSMUST00000118072.1 ENSMUST00000159721.1 |
Simc1
|
SUMO-interacting motifs containing 1 |
| chr1_-_87089928 | 1.69 |
ENSMUST00000027455.6
|
Alppl2
|
alkaline phosphatase, placental-like 2 |
| chr5_+_96209463 | 1.68 |
ENSMUST00000117766.1
|
Mrpl1
|
mitochondrial ribosomal protein L1 |
| chr16_-_38713235 | 1.66 |
ENSMUST00000023487.4
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr9_-_22131801 | 1.66 |
ENSMUST00000069330.6
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr9_+_48495345 | 1.63 |
ENSMUST00000048824.7
|
Gm5617
|
predicted gene 5617 |
| chr7_+_27607748 | 1.55 |
ENSMUST00000136962.1
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr4_+_53440388 | 1.52 |
ENSMUST00000102911.3
ENSMUST00000107646.2 |
Slc44a1
|
solute carrier family 44, member 1 |
| chr15_-_10470490 | 1.52 |
ENSMUST00000136591.1
|
Dnajc21
|
DnaJ (Hsp40) homolog, subfamily C, member 21 |
| chr1_+_132316112 | 1.51 |
ENSMUST00000082125.5
ENSMUST00000072177.7 |
Nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr6_+_83054653 | 1.46 |
ENSMUST00000092618.6
|
Aup1
|
ancient ubiquitous protein 1 |
| chr3_-_145649970 | 1.41 |
ENSMUST00000029846.3
|
Cyr61
|
cysteine rich protein 61 |
| chr5_-_108132577 | 1.37 |
ENSMUST00000061203.6
ENSMUST00000002837.4 |
Tmed5
|
transmembrane emp24 protein transport domain containing 5 |
| chr19_+_8840519 | 1.36 |
ENSMUST00000086058.6
|
Bscl2
|
Bernardinelli-Seip congenital lipodystrophy 2 homolog (human) |
| chr4_+_94614483 | 1.35 |
ENSMUST00000030311.4
ENSMUST00000107104.2 |
Ift74
|
intraflagellar transport 74 |
| chr4_+_116221590 | 1.31 |
ENSMUST00000147292.1
|
Pik3r3
|
phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 3 (p55) |
| chr2_-_91182848 | 1.28 |
ENSMUST00000111370.2
ENSMUST00000111376.1 ENSMUST00000099723.2 |
Madd
|
MAP-kinase activating death domain |
| chr2_+_54436317 | 1.26 |
ENSMUST00000112636.1
ENSMUST00000112635.1 ENSMUST00000112634.1 |
Galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr9_-_85749308 | 1.24 |
ENSMUST00000039213.8
|
Ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr1_-_172082757 | 1.23 |
ENSMUST00000003550.4
|
Ncstn
|
nicastrin |
| chr3_+_32436376 | 1.20 |
ENSMUST00000108242.1
|
Pik3ca
|
phosphatidylinositol 3-kinase, catalytic, alpha polypeptide |
| chr11_+_23306884 | 1.18 |
ENSMUST00000180046.1
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr2_+_52857844 | 1.17 |
ENSMUST00000090952.4
ENSMUST00000049483.6 ENSMUST00000050719.6 |
Fmnl2
|
formin-like 2 |
| chr1_-_172329261 | 1.16 |
ENSMUST00000062387.2
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr4_-_58785722 | 1.16 |
ENSMUST00000059608.2
|
Olfr267
|
olfactory receptor 267 |
| chr2_+_10047838 | 1.15 |
ENSMUST00000181588.1
|
C630004M23Rik
|
RIKEN cDNA C630004M23 gene |
| chr7_-_137314394 | 1.15 |
ENSMUST00000168203.1
ENSMUST00000106118.2 ENSMUST00000169486.2 ENSMUST00000033378.5 |
Ebf3
|
early B cell factor 3 |
| chr7_-_108930151 | 1.12 |
ENSMUST00000055745.3
|
Nlrp10
|
NLR family, pyrin domain containing 10 |
| chr1_+_170214826 | 1.12 |
ENSMUST00000159201.1
ENSMUST00000055830.1 |
4930500M09Rik
|
RIKEN cDNA 4930500M09 gene |
| chr4_+_116221633 | 1.11 |
ENSMUST00000030464.7
|
Pik3r3
|
phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 3 (p55) |
| chr9_+_54980880 | 1.10 |
ENSMUST00000093844.3
|
Chrna5
|
cholinergic receptor, nicotinic, alpha polypeptide 5 |
| chr10_+_90576252 | 1.08 |
ENSMUST00000182427.1
ENSMUST00000182053.1 ENSMUST00000182113.1 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr2_+_54436432 | 1.07 |
ENSMUST00000136642.1
|
Galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr11_+_58104229 | 1.05 |
ENSMUST00000108843.1
ENSMUST00000134896.1 |
Cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr17_+_36145018 | 1.05 |
ENSMUST00000173025.1
ENSMUST00000056774.9 |
2410017I17Rik
|
RIKEN cDNA 2410017I17 gene |
| chr18_-_60591783 | 1.04 |
ENSMUST00000056533.7
|
Myoz3
|
myozenin 3 |
| chr7_+_133637686 | 1.01 |
ENSMUST00000128901.2
|
2700050L05Rik
|
RIKEN cDNA 2700050L05 gene |
| chr19_+_4003334 | 1.01 |
ENSMUST00000025806.3
|
Doc2g
|
double C2, gamma |
| chr4_+_84884366 | 0.99 |
ENSMUST00000102819.3
|
Cntln
|
centlein, centrosomal protein |
| chr11_-_84068357 | 0.95 |
ENSMUST00000100705.4
|
Dusp14
|
dual specificity phosphatase 14 |
| chrX_+_152001845 | 0.95 |
ENSMUST00000026289.3
ENSMUST00000112617.3 |
Hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr17_+_46681038 | 0.94 |
ENSMUST00000002845.6
|
Mea1
|
male enhanced antigen 1 |
| chr11_-_23895208 | 0.93 |
ENSMUST00000102863.2
ENSMUST00000020513.3 |
Papolg
|
poly(A) polymerase gamma |
| chr13_+_40704005 | 0.92 |
ENSMUST00000069457.1
|
Gm9979
|
predicted gene 9979 |
| chr2_-_164828529 | 0.88 |
ENSMUST00000017911.3
|
Spata25
|
spermatogenesis associated 25 |
| chrX_+_42502596 | 0.85 |
ENSMUST00000115070.1
ENSMUST00000153948.1 |
Sh2d1a
|
SH2 domain protein 1A |
| chr6_-_142008745 | 0.84 |
ENSMUST00000111832.1
|
Gm6614
|
predicted gene 6614 |
| chr10_+_96136603 | 0.84 |
ENSMUST00000074615.6
|
Gm5426
|
predicted pseudogene 5426 |
| chr9_+_19641224 | 0.81 |
ENSMUST00000079042.6
|
Zfp317
|
zinc finger protein 317 |
| chr4_+_42950369 | 0.80 |
ENSMUST00000084662.5
|
Dnajb5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr12_-_31634592 | 0.79 |
ENSMUST00000020979.7
ENSMUST00000177962.1 |
Bcap29
|
B cell receptor associated protein 29 |
| chr1_+_172082509 | 0.78 |
ENSMUST00000135192.1
|
Copa
|
coatomer protein complex subunit alpha |
| chr15_-_99967067 | 0.77 |
ENSMUST00000100209.4
|
Fam186a
|
family with sequence similarity 186, member A |
| chr7_+_45486360 | 0.74 |
ENSMUST00000107758.1
|
Tulp2
|
tubby-like protein 2 |
| chr15_+_79229140 | 0.73 |
ENSMUST00000163571.1
|
Pick1
|
protein interacting with C kinase 1 |
| chr1_+_152954966 | 0.73 |
ENSMUST00000043313.8
|
Nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr7_-_30563184 | 0.71 |
ENSMUST00000043898.6
|
Psenen
|
presenilin enhancer 2 homolog (C. elegans) |
| chr1_+_172082796 | 0.69 |
ENSMUST00000027833.5
|
Copa
|
coatomer protein complex subunit alpha |
| chr6_+_15196949 | 0.67 |
ENSMUST00000151301.1
ENSMUST00000131414.1 ENSMUST00000140557.1 ENSMUST00000115469.1 |
Foxp2
|
forkhead box P2 |
| chr8_+_107119110 | 0.64 |
ENSMUST00000046116.1
|
C630050I24Rik
|
RIKEN cDNA C630050I24 gene |
| chr13_+_13590402 | 0.59 |
ENSMUST00000110559.1
|
Lyst
|
lysosomal trafficking regulator |
| chr2_+_164823001 | 0.56 |
ENSMUST00000132282.1
|
Zswim1
|
zinc finger SWIM-type containing 1 |
| chr6_+_42286676 | 0.56 |
ENSMUST00000031894.6
|
Clcn1
|
chloride channel 1 |
| chr4_+_42035113 | 0.55 |
ENSMUST00000098127.1
|
Gm10597
|
predicted gene 10597 |
| chr11_+_23306910 | 0.55 |
ENSMUST00000137823.1
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr6_+_42286709 | 0.54 |
ENSMUST00000163936.1
|
Clcn1
|
chloride channel 1 |
| chr16_+_19028232 | 0.44 |
ENSMUST00000074116.4
|
Gm10088
|
predicted gene 10088 |
| chr4_+_148000722 | 0.41 |
ENSMUST00000103230.4
|
Nppa
|
natriuretic peptide type A |
| chr4_+_116221689 | 0.37 |
ENSMUST00000106490.2
|
Pik3r3
|
phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 3 (p55) |
| chr2_+_68861564 | 0.35 |
ENSMUST00000176018.1
|
Cers6
|
ceramide synthase 6 |
| chr7_-_79793788 | 0.34 |
ENSMUST00000032760.5
|
Mesp1
|
mesoderm posterior 1 |
| chr1_-_71414881 | 0.33 |
ENSMUST00000087268.5
|
Abca12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr3_-_158562199 | 0.31 |
ENSMUST00000106044.1
|
Lrrc7
|
leucine rich repeat containing 7 |
| chr17_+_71552031 | 0.30 |
ENSMUST00000124001.1
ENSMUST00000167641.1 ENSMUST00000064420.5 |
Spdya
|
speedy homolog A (Xenopus laevis) |
| chr5_+_32136458 | 0.26 |
ENSMUST00000031017.9
|
Fosl2
|
fos-like antigen 2 |
| chr7_+_3645267 | 0.24 |
ENSMUST00000038913.9
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr2_+_152081529 | 0.22 |
ENSMUST00000064061.3
|
Scrt2
|
scratch homolog 2, zinc finger protein (Drosophila) |
| chr14_+_53616315 | 0.20 |
ENSMUST00000180972.1
ENSMUST00000185023.1 |
Trav12-2
|
T cell receptor alpha variable 12-2 |
| chr9_-_54647199 | 0.18 |
ENSMUST00000128163.1
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr4_+_84884276 | 0.17 |
ENSMUST00000047023.6
|
Cntln
|
centlein, centrosomal protein |
| chr3_+_96104498 | 0.16 |
ENSMUST00000132980.1
ENSMUST00000138206.1 ENSMUST00000090785.2 ENSMUST00000035519.5 |
Otud7b
|
OTU domain containing 7B |
| chr16_-_32868325 | 0.15 |
ENSMUST00000089684.3
ENSMUST00000040986.8 ENSMUST00000115105.2 |
1700021K19Rik
|
RIKEN cDNA 1700021K19 gene |
| chr7_+_105425817 | 0.15 |
ENSMUST00000181339.1
ENSMUST00000033189.4 |
Cckbr
|
cholecystokinin B receptor |
| chr13_-_103096818 | 0.14 |
ENSMUST00000166336.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr7_+_28071230 | 0.13 |
ENSMUST00000138392.1
ENSMUST00000076648.7 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr2_+_68861433 | 0.12 |
ENSMUST00000028426.2
|
Cers6
|
ceramide synthase 6 |
| chr8_-_93535707 | 0.09 |
ENSMUST00000077816.6
|
Ces5a
|
carboxylesterase 5A |
| chr4_-_141874879 | 0.09 |
ENSMUST00000036854.3
|
Efhd2
|
EF hand domain containing 2 |
| chr2_-_60673654 | 0.08 |
ENSMUST00000059888.8
ENSMUST00000154764.1 |
Itgb6
|
integrin beta 6 |
| chr1_-_173741717 | 0.07 |
ENSMUST00000127730.1
|
AI607873
|
expressed sequence AI607873 |
| chr10_-_128211788 | 0.05 |
ENSMUST00000061995.8
|
Spryd4
|
SPRY domain containing 4 |
| chrX_-_134474986 | 0.03 |
ENSMUST00000113223.2
|
Taf7l
|
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr10_+_101681487 | 0.03 |
ENSMUST00000179929.1
ENSMUST00000127504.1 |
Mgat4c
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr3_-_108154871 | 0.02 |
ENSMUST00000062028.1
|
Gpr61
|
G protein-coupled receptor 61 |
| chr4_+_114680769 | 0.02 |
ENSMUST00000146346.1
|
Gm12829
|
predicted gene 12829 |
| chr4_+_84884418 | 0.02 |
ENSMUST00000169371.2
|
Cntln
|
centlein, centrosomal protein |
| chrX_+_169685191 | 0.01 |
ENSMUST00000112104.1
ENSMUST00000112107.1 |
Mid1
|
midline 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.2 | 36.6 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 2.6 | 10.4 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 1.4 | 4.2 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 1.4 | 4.1 | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.3 | 5.0 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 1.1 | 17.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.9 | 2.8 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.9 | 3.6 | GO:0030091 | protein repair(GO:0030091) |
| 0.8 | 3.2 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.7 | 2.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.7 | 2.6 | GO:0061625 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.6 | 1.9 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.6 | 1.9 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.6 | 3.6 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.5 | 5.9 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.5 | 3.7 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.5 | 2.6 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.5 | 1.9 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.5 | 3.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.4 | 1.2 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.4 | 1.9 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.4 | 3.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.4 | 14.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.3 | 6.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.3 | 3.7 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 1.7 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.3 | 1.7 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.3 | 0.8 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 1.4 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.2 | 1.8 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.2 | 1.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 | 2.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.2 | 3.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 2.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 0.2 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 0.7 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 2.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.9 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 2.0 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 1.8 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.6 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 1.1 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.1 | 1.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 5.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.7 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 3.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 1.5 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.1 | 1.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.3 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 3.7 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 2.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 1.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.4 | GO:1903764 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 0.7 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 8.4 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.1 | 1.5 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.7 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 2.1 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 4.9 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 2.3 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 1.9 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 2.0 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 9.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 2.0 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 1.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 1.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 2.8 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 1.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 1.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.3 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.9 | 36.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.6 | 1.9 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.5 | 2.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.5 | 2.6 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.4 | 1.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.3 | 1.5 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.3 | 2.0 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 0.7 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.2 | 1.9 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 1.7 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.2 | 3.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 6.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 1.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.2 | 4.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 3.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 2.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 2.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 2.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 0.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 5.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 5.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 1.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 1.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 3.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.9 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 6.3 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.0 | 1.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 1.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 17.3 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 36.6 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 3.5 | 10.4 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.9 | 2.7 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.8 | 3.2 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.7 | 2.0 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.6 | 13.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.6 | 1.9 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.6 | 3.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.6 | 3.6 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.5 | 2.1 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.5 | 3.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.5 | 2.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.5 | 1.9 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.5 | 4.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.5 | 1.8 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.4 | 3.7 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.3 | 2.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.3 | 2.0 | GO:0005223 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.3 | 2.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 1.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 1.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 1.7 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.2 | 4.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 1.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 1.7 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 2.8 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.2 | 1.9 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 1.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 3.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 1.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 2.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 4.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 1.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.7 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 2.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 1.5 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 17.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.2 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.9 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 2.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.3 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.1 | 2.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 3.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.9 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 5.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 2.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 4.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 4.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 3.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 3.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.1 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 2.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 3.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 6.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 2.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 3.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.2 | GO:0017048 | Rho GTPase binding(GO:0017048) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 39.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 3.6 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 1.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 6.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.8 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 4.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 1.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 3.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 3.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.9 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.5 | 9.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.4 | 10.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.3 | 3.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 6.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 7.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 3.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 4.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 2.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 3.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 1.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.9 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 1.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 4.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.1 | 1.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 9.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 2.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 2.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 2.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 3.6 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |