Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Stat4_Stat3_Stat5b
Z-value: 0.90
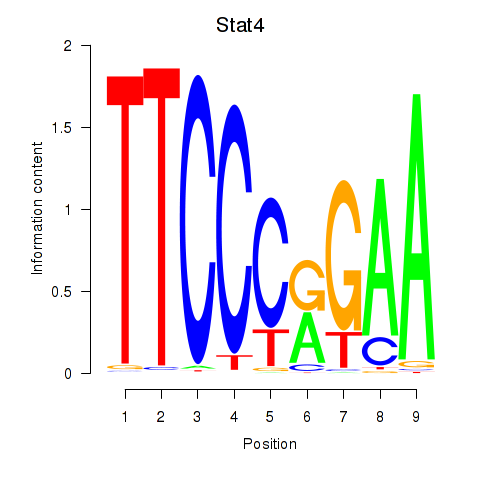
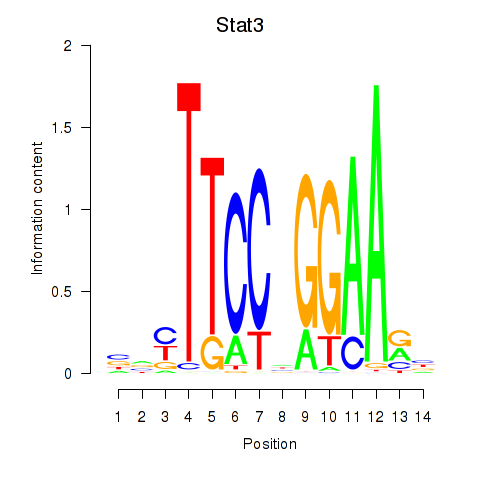
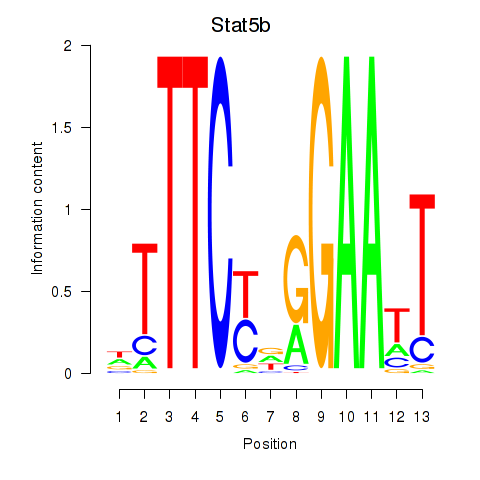
Transcription factors associated with Stat4_Stat3_Stat5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Stat4
|
ENSMUSG00000062939.5 | signal transducer and activator of transcription 4 |
|
Stat3
|
ENSMUSG00000004040.10 | signal transducer and activator of transcription 3 |
|
Stat5b
|
ENSMUSG00000020919.5 | signal transducer and activator of transcription 5B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat5b | mm10_v2_chr11_-_100822525_100822576 | 0.54 | 7.4e-04 | Click! |
| Stat3 | mm10_v2_chr11_-_100939540_100939566 | 0.40 | 1.6e-02 | Click! |
| Stat4 | mm10_v2_chr1_+_52008210_52008254 | -0.39 | 2.0e-02 | Click! |
Activity profile of Stat4_Stat3_Stat5b motif
Sorted Z-values of Stat4_Stat3_Stat5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_95417352 | 7.73 |
ENSMUST00000181781.1
|
5730420D15Rik
|
RIKEN cDNA 5730420D15 gene |
| chr14_+_30886476 | 6.54 |
ENSMUST00000006703.6
ENSMUST00000078490.5 ENSMUST00000120269.2 |
Itih4
|
inter alpha-trypsin inhibitor, heavy chain 4 |
| chr14_+_30886521 | 6.52 |
ENSMUST00000168782.1
|
Itih4
|
inter alpha-trypsin inhibitor, heavy chain 4 |
| chr10_-_95417099 | 5.37 |
ENSMUST00000135822.1
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr8_+_105269837 | 5.29 |
ENSMUST00000172525.1
ENSMUST00000174837.1 ENSMUST00000173859.1 |
Hsf4
|
heat shock transcription factor 4 |
| chr10_-_95416850 | 5.09 |
ENSMUST00000020215.9
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr7_+_107567445 | 4.33 |
ENSMUST00000120990.1
|
Olfml1
|
olfactomedin-like 1 |
| chr9_+_107296843 | 4.02 |
ENSMUST00000167072.1
|
Cish
|
cytokine inducible SH2-containing protein |
| chr8_-_124569696 | 3.78 |
ENSMUST00000063278.6
|
Agt
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
| chr11_+_70054334 | 3.53 |
ENSMUST00000018699.6
ENSMUST00000108585.2 |
Asgr1
|
asialoglycoprotein receptor 1 |
| chr5_+_7960445 | 3.46 |
ENSMUST00000115421.1
|
Steap4
|
STEAP family member 4 |
| chr17_-_56121946 | 3.14 |
ENSMUST00000041357.7
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr16_+_62854299 | 3.10 |
ENSMUST00000023629.8
|
Pros1
|
protein S (alpha) |
| chr13_-_41847482 | 3.06 |
ENSMUST00000072012.3
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr13_-_41847626 | 3.01 |
ENSMUST00000121404.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr9_+_107296682 | 2.98 |
ENSMUST00000168260.1
|
Cish
|
cytokine inducible SH2-containing protein |
| chr11_-_83530505 | 2.84 |
ENSMUST00000035938.2
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr9_-_45009590 | 2.79 |
ENSMUST00000102832.1
|
Cd3e
|
CD3 antigen, epsilon polypeptide |
| chr13_-_41847599 | 2.78 |
ENSMUST00000179758.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chrX_-_139782353 | 2.70 |
ENSMUST00000101217.3
|
Ripply1
|
ripply1 homolog (zebrafish) |
| chrX_-_108664891 | 2.68 |
ENSMUST00000178160.1
|
Gm379
|
predicted gene 379 |
| chr15_-_5063741 | 2.62 |
ENSMUST00000110689.3
|
C7
|
complement component 7 |
| chr7_-_81706905 | 2.61 |
ENSMUST00000026922.7
|
Homer2
|
homer homolog 2 (Drosophila) |
| chrX_+_59999436 | 2.60 |
ENSMUST00000033477.4
|
F9
|
coagulation factor IX |
| chr18_-_3281036 | 2.60 |
ENSMUST00000049942.6
ENSMUST00000139537.1 ENSMUST00000124747.1 |
Crem
|
cAMP responsive element modulator |
| chr15_+_10223974 | 2.53 |
ENSMUST00000128450.1
ENSMUST00000148257.1 ENSMUST00000128921.1 |
Prlr
|
prolactin receptor |
| chr10_-_109010955 | 2.51 |
ENSMUST00000105276.1
ENSMUST00000064054.7 |
Syt1
|
synaptotagmin I |
| chr6_+_41521782 | 2.50 |
ENSMUST00000070380.4
|
Prss2
|
protease, serine, 2 |
| chr7_-_30924169 | 2.47 |
ENSMUST00000074671.6
|
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr10_-_127070254 | 2.27 |
ENSMUST00000060991.4
|
Tspan31
|
tetraspanin 31 |
| chr11_+_49087022 | 2.19 |
ENSMUST00000046704.6
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr11_+_78499087 | 2.17 |
ENSMUST00000017488.4
|
Vtn
|
vitronectin |
| chr3_+_19957088 | 2.15 |
ENSMUST00000108328.1
|
Cp
|
ceruloplasmin |
| chr4_+_148602527 | 2.13 |
ENSMUST00000105701.2
ENSMUST00000052060.6 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr15_-_37458523 | 2.09 |
ENSMUST00000116445.2
|
Ncald
|
neurocalcin delta |
| chr15_-_60921270 | 2.05 |
ENSMUST00000096418.3
|
A1bg
|
alpha-1-B glycoprotein |
| chr5_+_130448801 | 2.03 |
ENSMUST00000111288.2
|
Caln1
|
calneuron 1 |
| chr16_-_35871544 | 2.03 |
ENSMUST00000042665.8
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr3_+_19957037 | 2.00 |
ENSMUST00000091309.5
ENSMUST00000108329.1 ENSMUST00000003714.6 |
Cp
|
ceruloplasmin |
| chr7_+_26808880 | 1.90 |
ENSMUST00000040944.7
|
Cyp2g1
|
cytochrome P450, family 2, subfamily g, polypeptide 1 |
| chr8_+_104926237 | 1.88 |
ENSMUST00000034355.4
ENSMUST00000109410.2 |
Ces2e
|
carboxylesterase 2E |
| chr3_+_19957240 | 1.82 |
ENSMUST00000108325.2
|
Cp
|
ceruloplasmin |
| chr1_+_171214013 | 1.77 |
ENSMUST00000111328.1
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr19_+_38481057 | 1.75 |
ENSMUST00000182481.1
|
Plce1
|
phospholipase C, epsilon 1 |
| chr4_-_140617062 | 1.75 |
ENSMUST00000154979.1
|
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr1_-_169747634 | 1.73 |
ENSMUST00000027991.5
ENSMUST00000111357.1 |
Rgs4
|
regulator of G-protein signaling 4 |
| chr4_-_108031938 | 1.73 |
ENSMUST00000106708.1
|
Podn
|
podocan |
| chr4_-_108032069 | 1.70 |
ENSMUST00000106709.2
|
Podn
|
podocan |
| chr19_+_4711153 | 1.66 |
ENSMUST00000008991.6
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr1_-_155812805 | 1.65 |
ENSMUST00000111764.2
|
Qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr9_+_124021965 | 1.64 |
ENSMUST00000039171.7
|
Ccr3
|
chemokine (C-C motif) receptor 3 |
| chr12_-_103989950 | 1.64 |
ENSMUST00000120251.2
|
Serpina11
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
| chr1_-_155812859 | 1.64 |
ENSMUST00000035325.8
|
Qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr9_-_105960642 | 1.63 |
ENSMUST00000165165.2
|
Col6a5
|
collagen, type VI, alpha 5 |
| chr7_-_141276729 | 1.62 |
ENSMUST00000167263.1
ENSMUST00000080654.5 |
Cdhr5
|
cadherin-related family member 5 |
| chr7_+_28180272 | 1.57 |
ENSMUST00000173223.1
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr12_-_103989917 | 1.50 |
ENSMUST00000151709.2
ENSMUST00000176246.1 ENSMUST00000074693.7 |
Serpina11
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
| chr14_-_55713482 | 1.49 |
ENSMUST00000168729.1
ENSMUST00000178034.1 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr11_+_82035569 | 1.47 |
ENSMUST00000000193.5
|
Ccl2
|
chemokine (C-C motif) ligand 2 |
| chr9_-_71163224 | 1.47 |
ENSMUST00000074465.2
|
Aqp9
|
aquaporin 9 |
| chr7_+_28180226 | 1.44 |
ENSMUST00000172467.1
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr6_+_121346618 | 1.42 |
ENSMUST00000032200.9
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr6_+_41354105 | 1.41 |
ENSMUST00000072103.5
|
Try10
|
trypsin 10 |
| chr1_+_171213969 | 1.39 |
ENSMUST00000005820.4
ENSMUST00000075469.5 ENSMUST00000155126.1 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr17_-_34187219 | 1.34 |
ENSMUST00000173831.1
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr6_-_41314700 | 1.33 |
ENSMUST00000064324.5
|
Try5
|
trypsin 5 |
| chr5_+_88886809 | 1.31 |
ENSMUST00000148750.1
|
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4 |
| chr17_+_34187545 | 1.31 |
ENSMUST00000170086.1
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr5_+_90561102 | 1.29 |
ENSMUST00000094615.4
|
5830473C10Rik
|
RIKEN cDNA 5830473C10 gene |
| chr15_-_54919961 | 1.28 |
ENSMUST00000167541.2
ENSMUST00000041591.9 ENSMUST00000173516.1 |
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr15_-_54920115 | 1.26 |
ENSMUST00000171545.1
|
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr16_-_23988852 | 1.25 |
ENSMUST00000023151.5
|
Bcl6
|
B cell leukemia/lymphoma 6 |
| chr17_+_43667389 | 1.24 |
ENSMUST00000170988.1
|
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr17_+_34187789 | 1.22 |
ENSMUST00000041633.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr11_-_48994172 | 1.19 |
ENSMUST00000146439.1
|
Tgtp1
|
T cell specific GTPase 1 |
| chr14_+_65970610 | 1.19 |
ENSMUST00000127387.1
|
Clu
|
clusterin |
| chr14_+_65970804 | 1.17 |
ENSMUST00000138191.1
|
Clu
|
clusterin |
| chr4_+_11704439 | 1.17 |
ENSMUST00000108304.2
|
Gem
|
GTP binding protein (gene overexpressed in skeletal muscle) |
| chr17_-_78985428 | 1.14 |
ENSMUST00000118991.1
|
Prkd3
|
protein kinase D3 |
| chr8_-_110805863 | 1.12 |
ENSMUST00000150680.1
ENSMUST00000076846.4 |
Il34
|
interleukin 34 |
| chr14_+_65971049 | 1.12 |
ENSMUST00000128539.1
|
Clu
|
clusterin |
| chr9_-_51328898 | 1.12 |
ENSMUST00000039959.4
|
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr7_+_127800604 | 1.11 |
ENSMUST00000046863.5
ENSMUST00000106272.1 ENSMUST00000139068.1 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr2_-_84775420 | 1.11 |
ENSMUST00000111641.1
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr1_+_133363564 | 1.11 |
ENSMUST00000135222.2
|
Etnk2
|
ethanolamine kinase 2 |
| chr1_-_170976112 | 1.09 |
ENSMUST00000027966.7
ENSMUST00000081103.5 ENSMUST00000159688.1 |
Fcgr2b
|
Fc receptor, IgG, low affinity IIb |
| chr2_-_84775388 | 1.09 |
ENSMUST00000023994.3
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr7_-_105600103 | 1.07 |
ENSMUST00000033185.8
|
Hpx
|
hemopexin |
| chr6_+_34598500 | 1.06 |
ENSMUST00000079391.3
ENSMUST00000142512.1 |
Cald1
|
caldesmon 1 |
| chr15_+_102102926 | 1.06 |
ENSMUST00000169627.1
ENSMUST00000046144.9 |
Tenc1
|
tensin like C1 domain-containing phosphatase |
| chr9_-_107679592 | 1.06 |
ENSMUST00000010205.7
|
Gnat1
|
guanine nucleotide binding protein, alpha transducing 1 |
| chr4_-_57916283 | 1.04 |
ENSMUST00000063816.5
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene |
| chr14_+_65971164 | 1.03 |
ENSMUST00000144619.1
|
Clu
|
clusterin |
| chr6_+_138141569 | 1.02 |
ENSMUST00000118091.1
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr6_+_34598530 | 1.01 |
ENSMUST00000115027.1
ENSMUST00000115026.1 |
Cald1
|
caldesmon 1 |
| chr15_-_37459327 | 1.00 |
ENSMUST00000119730.1
ENSMUST00000120746.1 |
Ncald
|
neurocalcin delta |
| chr14_-_56262233 | 0.99 |
ENSMUST00000015581.4
|
Gzmb
|
granzyme B |
| chr7_+_127800844 | 0.99 |
ENSMUST00000106271.1
ENSMUST00000138432.1 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr3_+_142496924 | 0.97 |
ENSMUST00000090127.2
|
Gbp5
|
guanylate binding protein 5 |
| chr4_-_49473905 | 0.97 |
ENSMUST00000135976.1
|
Acnat1
|
acyl-coenzyme A amino acid N-acyltransferase 1 |
| chr4_-_116017854 | 0.96 |
ENSMUST00000049095.5
|
Faah
|
fatty acid amide hydrolase |
| chr9_+_21015960 | 0.96 |
ENSMUST00000086399.4
|
Icam1
|
intercellular adhesion molecule 1 |
| chr15_-_89170688 | 0.94 |
ENSMUST00000060808.9
|
Plxnb2
|
plexin B2 |
| chr16_+_38902305 | 0.91 |
ENSMUST00000023478.7
|
Igsf11
|
immunoglobulin superfamily, member 11 |
| chr19_-_21652714 | 0.86 |
ENSMUST00000177577.1
|
1110059E24Rik
|
RIKEN cDNA 1110059E24 gene |
| chr19_-_29047847 | 0.85 |
ENSMUST00000025696.4
|
Ak3
|
adenylate kinase 3 |
| chr11_-_82764303 | 0.85 |
ENSMUST00000021040.3
ENSMUST00000100722.4 |
Cct6b
|
chaperonin containing Tcp1, subunit 6b (zeta) |
| chr5_-_66151323 | 0.85 |
ENSMUST00000131838.1
|
Rbm47
|
RNA binding motif protein 47 |
| chr2_-_101628958 | 0.85 |
ENSMUST00000111231.3
|
B230118H07Rik
|
RIKEN cDNA B230118H07 gene |
| chr9_-_44526397 | 0.84 |
ENSMUST00000062215.7
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr1_+_136624901 | 0.84 |
ENSMUST00000047734.8
ENSMUST00000112046.1 |
Zfp281
|
zinc finger protein 281 |
| chr2_-_101628930 | 0.83 |
ENSMUST00000099682.2
|
B230118H07Rik
|
RIKEN cDNA B230118H07 gene |
| chr3_+_89229046 | 0.83 |
ENSMUST00000041142.3
|
Muc1
|
mucin 1, transmembrane |
| chr17_-_32420965 | 0.82 |
ENSMUST00000170392.1
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr1_-_172895048 | 0.81 |
ENSMUST00000027824.5
|
Apcs
|
serum amyloid P-component |
| chr8_+_13895816 | 0.80 |
ENSMUST00000084055.7
|
Gm7676
|
predicted gene 7676 |
| chr12_+_88953399 | 0.80 |
ENSMUST00000057634.7
|
Nrxn3
|
neurexin III |
| chr1_-_9298499 | 0.79 |
ENSMUST00000132064.1
|
Sntg1
|
syntrophin, gamma 1 |
| chr14_-_55713088 | 0.78 |
ENSMUST00000002389.7
|
Tgm1
|
transglutaminase 1, K polypeptide |
| chr17_-_74047850 | 0.78 |
ENSMUST00000043458.7
|
Srd5a2
|
steroid 5 alpha-reductase 2 |
| chr9_-_106476104 | 0.78 |
ENSMUST00000156426.1
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr8_+_94532990 | 0.77 |
ENSMUST00000048653.2
ENSMUST00000109537.1 |
Cpne2
|
copine II |
| chr3_+_63295815 | 0.77 |
ENSMUST00000029400.1
|
Mme
|
membrane metallo endopeptidase |
| chr11_-_60046477 | 0.77 |
ENSMUST00000000310.7
ENSMUST00000102693.2 ENSMUST00000148512.1 |
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr3_-_98753465 | 0.76 |
ENSMUST00000094050.4
ENSMUST00000090743.6 |
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
| chr2_-_168712853 | 0.76 |
ENSMUST00000123156.1
ENSMUST00000156555.1 |
Atp9a
|
ATPase, class II, type 9A |
| chr7_+_24134148 | 0.76 |
ENSMUST00000056549.7
|
Zfp235
|
zinc finger protein 235 |
| chr7_-_105399991 | 0.74 |
ENSMUST00000118726.1
ENSMUST00000074686.7 ENSMUST00000122327.1 ENSMUST00000179474.1 ENSMUST00000048079.6 |
Fam160a2
|
family with sequence similarity 160, member A2 |
| chr16_+_90738324 | 0.74 |
ENSMUST00000038197.2
|
Mrap
|
melanocortin 2 receptor accessory protein |
| chr12_-_45074112 | 0.74 |
ENSMUST00000120531.1
ENSMUST00000143376.1 |
Stxbp6
|
syntaxin binding protein 6 (amisyn) |
| chr8_+_105326354 | 0.73 |
ENSMUST00000015000.5
ENSMUST00000098453.2 |
Tmem208
|
transmembrane protein 208 |
| chr10_-_95415283 | 0.73 |
ENSMUST00000119917.1
|
Socs2
|
suppressor of cytokine signaling 2 |
| chrX_+_57212110 | 0.73 |
ENSMUST00000033466.1
|
Cd40lg
|
CD40 ligand |
| chr10_+_13966268 | 0.73 |
ENSMUST00000015645.4
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr6_-_124911636 | 0.72 |
ENSMUST00000032217.1
|
Lag3
|
lymphocyte-activation gene 3 |
| chr7_-_30944017 | 0.72 |
ENSMUST00000062620.7
|
Hamp
|
hepcidin antimicrobial peptide |
| chr3_+_122895072 | 0.71 |
ENSMUST00000023820.5
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr16_-_30267524 | 0.70 |
ENSMUST00000064856.7
|
Cpn2
|
carboxypeptidase N, polypeptide 2 |
| chr7_+_24270420 | 0.70 |
ENSMUST00000108438.3
|
Zfp93
|
zinc finger protein 93 |
| chr2_-_77170534 | 0.70 |
ENSMUST00000111833.2
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr17_+_35470083 | 0.69 |
ENSMUST00000174525.1
ENSMUST00000068291.6 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr7_-_24208093 | 0.69 |
ENSMUST00000086006.5
|
Zfp111
|
zinc finger protein 111 |
| chr18_-_33463615 | 0.69 |
ENSMUST00000051087.8
|
Nrep
|
neuronal regeneration related protein |
| chr1_-_171240055 | 0.69 |
ENSMUST00000131286.1
|
Ndufs2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2 |
| chr11_+_58215028 | 0.68 |
ENSMUST00000108836.1
|
Irgm2
|
immunity-related GTPase family M member 2 |
| chr6_+_108660772 | 0.68 |
ENSMUST00000163617.1
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr2_-_120314141 | 0.66 |
ENSMUST00000054651.7
|
Pla2g4f
|
phospholipase A2, group IVF |
| chr15_+_100304782 | 0.66 |
ENSMUST00000067752.3
|
Mettl7a1
|
methyltransferase like 7A1 |
| chr19_-_4477447 | 0.65 |
ENSMUST00000059295.3
|
Syt12
|
synaptotagmin XII |
| chr13_+_112464070 | 0.65 |
ENSMUST00000183663.1
ENSMUST00000184311.1 ENSMUST00000183886.1 |
Il6st
|
interleukin 6 signal transducer |
| chr3_+_83007850 | 0.64 |
ENSMUST00000048486.7
|
Fgg
|
fibrinogen gamma chain |
| chr6_-_18030435 | 0.64 |
ENSMUST00000010941.2
|
Wnt2
|
wingless-related MMTV integration site 2 |
| chr11_-_21572193 | 0.64 |
ENSMUST00000102874.4
|
Mdh1
|
malate dehydrogenase 1, NAD (soluble) |
| chr19_-_58455161 | 0.63 |
ENSMUST00000135730.1
ENSMUST00000152507.1 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr15_+_89076124 | 0.63 |
ENSMUST00000165690.1
|
Trabd
|
TraB domain containing |
| chr11_-_113710017 | 0.62 |
ENSMUST00000018871.1
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr1_+_71652837 | 0.62 |
ENSMUST00000097699.2
|
Apol7d
|
apolipoprotein L 7d |
| chr7_+_102210335 | 0.62 |
ENSMUST00000140631.1
ENSMUST00000120879.1 ENSMUST00000146996.1 |
Pgap2
|
post-GPI attachment to proteins 2 |
| chr11_+_120530688 | 0.61 |
ENSMUST00000026119.7
|
Gcgr
|
glucagon receptor |
| chr11_+_53770014 | 0.61 |
ENSMUST00000108920.2
ENSMUST00000140866.1 ENSMUST00000108922.1 |
Irf1
|
interferon regulatory factor 1 |
| chr11_-_69695802 | 0.61 |
ENSMUST00000108649.1
ENSMUST00000174159.1 ENSMUST00000181810.1 |
BC096441
Tnfsf12
|
cDNA sequence BC096441 tumor necrosis factor (ligand) superfamily, member 12 |
| chr17_-_28350747 | 0.61 |
ENSMUST00000080572.7
ENSMUST00000156862.1 |
Tead3
|
TEA domain family member 3 |
| chr11_-_49113757 | 0.60 |
ENSMUST00000060398.1
|
Olfr1396
|
olfactory receptor 1396 |
| chr11_-_106579111 | 0.59 |
ENSMUST00000103070.2
|
Tex2
|
testis expressed gene 2 |
| chr6_-_122282810 | 0.59 |
ENSMUST00000032207.8
|
Klrg1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr11_-_53430417 | 0.59 |
ENSMUST00000109019.1
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr15_+_100353149 | 0.59 |
ENSMUST00000075675.5
ENSMUST00000088142.5 ENSMUST00000176287.1 |
AB099516
Mettl7a2
|
cDNA sequence AB099516 methyltransferase like 7A2 |
| chr9_-_103230415 | 0.59 |
ENSMUST00000035158.9
|
Trf
|
transferrin |
| chr1_+_164796723 | 0.59 |
ENSMUST00000027861.4
|
Dpt
|
dermatopontin |
| chr10_-_54075702 | 0.58 |
ENSMUST00000105470.1
|
Man1a
|
mannosidase 1, alpha |
| chr17_+_35126316 | 0.58 |
ENSMUST00000061859.6
|
D17H6S53E
|
DNA segment, Chr 17, human D6S53E |
| chr16_-_24393588 | 0.58 |
ENSMUST00000181640.1
|
1110054M08Rik
|
RIKEN cDNA 1110054M08 gene |
| chr4_-_108118528 | 0.58 |
ENSMUST00000030340.8
|
Scp2
|
sterol carrier protein 2, liver |
| chr5_-_66150898 | 0.57 |
ENSMUST00000113725.1
ENSMUST00000094757.2 |
Rbm47
|
RNA binding motif protein 47 |
| chr13_-_17694729 | 0.57 |
ENSMUST00000068545.4
|
5033411D12Rik
|
RIKEN cDNA 5033411D12 gene |
| chr11_-_69695753 | 0.57 |
ENSMUST00000180946.1
|
Tnfsf12
|
tumor necrosis factor (ligand) superfamily, member 12 |
| chr11_-_51857624 | 0.57 |
ENSMUST00000020655.7
ENSMUST00000109090.1 |
Phf15
|
PHD finger protein 15 |
| chr2_+_101886249 | 0.57 |
ENSMUST00000028584.7
|
Commd9
|
COMM domain containing 9 |
| chr9_+_54286479 | 0.57 |
ENSMUST00000056740.5
|
Gldn
|
gliomedin |
| chr8_+_107150621 | 0.57 |
ENSMUST00000034400.3
|
Cyb5b
|
cytochrome b5 type B |
| chr7_-_127051948 | 0.57 |
ENSMUST00000051122.5
|
Zg16
|
zymogen granule protein 16 |
| chr7_-_99182681 | 0.57 |
ENSMUST00000033001.4
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr9_+_20868628 | 0.57 |
ENSMUST00000043911.7
|
A230050P20Rik
|
RIKEN cDNA A230050P20 gene |
| chr11_-_69695521 | 0.56 |
ENSMUST00000181261.1
|
Tnfsf12
|
tumor necrosis factor (ligand) superfamily, member 12 |
| chr5_-_103977360 | 0.56 |
ENSMUST00000048118.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr10_+_18235030 | 0.56 |
ENSMUST00000181897.1
|
Gm10827
|
predicted gene 10827 |
| chr5_-_103977326 | 0.56 |
ENSMUST00000120320.1
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr17_+_56628118 | 0.56 |
ENSMUST00000112979.2
|
Catsperd
|
catsper channel auxiliary subunit delta |
| chr7_+_38183787 | 0.56 |
ENSMUST00000067854.8
ENSMUST00000177983.1 |
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene |
| chr10_-_54075730 | 0.56 |
ENSMUST00000105469.1
ENSMUST00000003843.8 |
Man1a
|
mannosidase 1, alpha |
| chr14_+_27622433 | 0.56 |
ENSMUST00000090302.5
|
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr7_-_78577771 | 0.55 |
ENSMUST00000039438.7
|
Ntrk3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr7_+_28179469 | 0.55 |
ENSMUST00000085901.6
ENSMUST00000172761.1 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr12_+_28675220 | 0.55 |
ENSMUST00000020957.6
|
Adi1
|
acireductone dioxygenase 1 |
| chr7_+_30095150 | 0.55 |
ENSMUST00000130526.1
ENSMUST00000108200.1 |
Zfp260
|
zinc finger protein 260 |
| chr4_+_134396320 | 0.55 |
ENSMUST00000105869.2
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr3_-_85741389 | 0.55 |
ENSMUST00000094148.4
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr5_+_35041539 | 0.55 |
ENSMUST00000030985.6
|
Hgfac
|
hepatocyte growth factor activator |
| chr5_-_103977404 | 0.54 |
ENSMUST00000112803.2
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr8_+_127447669 | 0.54 |
ENSMUST00000159511.1
|
Pard3
|
par-3 (partitioning defective 3) homolog (C. elegans) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Stat4_Stat3_Stat5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) L-lysine transport(GO:1902022) |
| 0.8 | 3.2 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.8 | 0.8 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.8 | 4.5 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.7 | 2.2 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.7 | 2.8 | GO:2000503 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.6 | 2.6 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.6 | 1.2 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.6 | 2.8 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.5 | 2.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.5 | 2.5 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.5 | 2.8 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.5 | 11.6 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.4 | 1.2 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.4 | 1.1 | GO:1902605 | heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.4 | 1.5 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.4 | 2.5 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.4 | 1.4 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.3 | 3.5 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.3 | 13.0 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.3 | 1.6 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.3 | 2.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 2.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.3 | 2.7 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.3 | 1.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 0.8 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.3 | 2.9 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 0.8 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.3 | 1.0 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.2 | 0.7 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.2 | 6.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 1.6 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.2 | 5.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 2.4 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.2 | 0.6 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.2 | 0.6 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.2 | 1.1 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) |
| 0.2 | 1.0 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.2 | 2.2 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.2 | 1.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.2 | 0.5 | GO:2000314 | negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.2 | 0.7 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.2 | 2.7 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.2 | 0.9 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.2 | 0.3 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.2 | 4.0 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.2 | 1.0 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.2 | 0.5 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.2 | 1.6 | GO:0002551 | mast cell chemotaxis(GO:0002551) mast cell migration(GO:0097531) |
| 0.2 | 1.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 0.8 | GO:0032824 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.2 | 0.5 | GO:0015755 | fructose transport(GO:0015755) cellular response to fructose stimulus(GO:0071332) |
| 0.2 | 0.9 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.2 | 2.6 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.2 | 0.3 | GO:0071724 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.2 | 0.6 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.2 | 0.9 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 1.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.6 | GO:0032385 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.1 | 0.9 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 5.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.6 | GO:0048691 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.1 | 0.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.6 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 2.6 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 1.9 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.4 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 1.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 1.0 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 0.7 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.7 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.1 | 3.4 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 0.3 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.1 | 0.5 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.4 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 | 0.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.4 | GO:1903027 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.2 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 | 0.5 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.1 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.1 | 1.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.1 | 0.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.3 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.1 | 0.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.5 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.4 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.8 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 1.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 3.1 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 3.5 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 0.8 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.6 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 0.3 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 0.6 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.1 | 0.2 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) |
| 0.1 | 0.2 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 0.4 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.5 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.3 | GO:0000436 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) carbon catabolite activation of transcription(GO:0045991) |
| 0.1 | 0.5 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.2 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.1 | 1.0 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 0.2 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 1.7 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.4 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 0.2 | GO:0035627 | ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.1 | 0.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.8 | GO:0042448 | progesterone metabolic process(GO:0042448) testosterone biosynthetic process(GO:0061370) |
| 0.1 | 3.6 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.2 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.1 | 0.3 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 0.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.3 | GO:2001182 | regulation of interleukin-12 secretion(GO:2001182) |
| 0.1 | 0.3 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.1 | 0.3 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.7 | GO:0071236 | cellular response to antibiotic(GO:0071236) |
| 0.1 | 0.8 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.2 | GO:0032242 | positive regulation of necrotic cell death(GO:0010940) regulation of nucleoside transport(GO:0032242) |
| 0.1 | 1.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.2 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 1.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 0.4 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.3 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.6 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.5 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.6 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.4 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.3 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.2 | GO:0021666 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.0 | 1.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.1 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.2 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.4 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 1.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.2 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.0 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.2 | GO:0070164 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.8 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.5 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.4 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 2.8 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.4 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.8 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.3 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.0 | 1.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 1.4 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.3 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) |
| 0.0 | 0.3 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.2 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 2.0 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.3 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.0 | 0.1 | GO:0060995 | pro-T cell differentiation(GO:0002572) cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.0 | 1.5 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.1 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.0 | 0.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0072530 | purine-containing compound transmembrane transport(GO:0072530) |
| 0.0 | 0.7 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.1 | GO:1903011 | negative regulation of bone development(GO:1903011) negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 3.2 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) termination of signal transduction(GO:0023021) |
| 0.0 | 0.5 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) negative regulation of biomineral tissue development(GO:0070168) |
| 0.0 | 2.9 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.2 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 1.1 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.0 | 1.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:0032899 | regulation of neurotrophin production(GO:0032899) positive regulation of neurotrophin production(GO:0032901) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.3 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.2 | GO:0097647 | basement membrane assembly(GO:0070831) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.7 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.0 | 1.4 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.2 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 1.8 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.6 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.0 | 0.5 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 2.0 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 0.9 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.6 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.4 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:2000321 | negative regulation of interleukin-1 beta secretion(GO:0050713) positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.1 | GO:0015791 | polyol transport(GO:0015791) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.6 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 3.0 | GO:0031668 | cellular response to extracellular stimulus(GO:0031668) |
| 0.0 | 0.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.5 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.5 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 2.0 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 0.1 | GO:0033561 | regulation of water loss via skin(GO:0033561) establishment of skin barrier(GO:0061436) |
| 0.0 | 0.0 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 2.6 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.1 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.2 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.4 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 0.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.8 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0044299 | C-fiber(GO:0044299) |
| 0.4 | 2.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.4 | 3.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.3 | 4.5 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.3 | 2.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.3 | 1.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 2.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 2.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 1.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 35.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 0.6 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.2 | 1.0 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.2 | 0.8 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.2 | 0.5 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 1.6 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 2.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.4 | GO:0036019 | endolysosome(GO:0036019) |
| 0.1 | 1.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 2.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.7 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.9 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.2 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.4 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 0.9 | GO:0005767 | secondary lysosome(GO:0005767) autolysosome(GO:0044754) |
| 0.1 | 0.6 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 3.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.2 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 2.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.3 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.4 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.7 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.1 | 0.7 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.5 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 1.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 2.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.8 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 1.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.2 | GO:0097342 | death-inducing signaling complex(GO:0031264) ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 1.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 11.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.9 | 2.8 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.8 | 3.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.7 | 2.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.6 | 2.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.6 | 6.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.5 | 12.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.5 | 2.5 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.5 | 2.5 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.4 | 3.5 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.4 | 2.5 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.3 | 3.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.3 | 1.5 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.3 | 0.9 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.3 | 2.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 1.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.3 | 0.8 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.3 | 1.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 1.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 1.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 3.8 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.2 | 0.6 | GO:0004923 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.2 | 0.6 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 0.6 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.2 | 2.9 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 0.6 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.2 | 5.2 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 0.8 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.2 | 0.6 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.2 | 4.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 1.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.2 | 1.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 1.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 0.5 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.2 | 0.8 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.9 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 1.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.6 | GO:1904121 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.1 | 1.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 2.0 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.3 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.1 | 0.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.9 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 1.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.1 | GO:0046977 | TAP binding(GO:0046977) |
| 0.1 | 0.4 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.8 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.6 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 2.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.6 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 1.3 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 0.4 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 3.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.4 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 0.6 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 9.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.3 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 1.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.2 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.1 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.2 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.1 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.4 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.3 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.6 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.1 | 3.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 3.7 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.1 | 0.7 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.9 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.5 | GO:0070061 | fructose binding(GO:0070061) |
| 0.1 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) lipopeptide binding(GO:0071723) |
| 0.0 | 1.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 8.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.7 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 1.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 1.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.6 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 1.0 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.2 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.3 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.3 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.2 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 6.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 2.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.3 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 1.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.1 | GO:0019962 | interferon receptor activity(GO:0004904) type I interferon receptor activity(GO:0004905) type I interferon binding(GO:0019962) |
| 0.0 | 0.2 | GO:0032813 | death receptor binding(GO:0005123) tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 2.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 1.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0043531 | ADP binding(GO:0043531) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 4.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 12.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 4.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 26.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 7.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 3.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 2.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 4.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 2.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 3.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 2.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 21.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.4 | 5.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 3.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.3 | 2.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 3.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 2.5 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.2 | 2.5 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 6.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 6.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 2.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.4 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 1.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 2.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 0.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 2.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 0.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 1.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.6 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 3.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 7.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 2.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.8 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.0 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.7 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |