Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
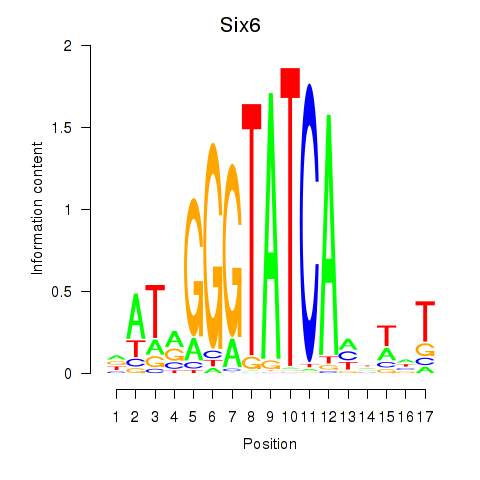
Results for Six6
Z-value: 1.96
Transcription factors associated with Six6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Six6
|
ENSMUSG00000021099.5 | sine oculis-related homeobox 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Six6 | mm10_v2_chr12_+_72939724_72939758 | 0.25 | 1.3e-01 | Click! |
Activity profile of Six6 motif
Sorted Z-values of Six6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_117797660 | 25.87 |
ENSMUST00000106331.1
|
6030468B19Rik
|
RIKEN cDNA 6030468B19 gene |
| chr4_+_134864536 | 24.45 |
ENSMUST00000030627.7
|
Rhd
|
Rh blood group, D antigen |
| chr17_+_48247759 | 20.75 |
ENSMUST00000048065.5
|
Trem3
|
triggering receptor expressed on myeloid cells 3 |
| chr6_-_41314700 | 19.80 |
ENSMUST00000064324.5
|
Try5
|
trypsin 5 |
| chr17_-_28560704 | 19.65 |
ENSMUST00000114785.1
ENSMUST00000025062.3 |
Clps
|
colipase, pancreatic |
| chr16_-_36408349 | 19.38 |
ENSMUST00000023619.6
|
Stfa2
|
stefin A2 |
| chr6_+_41458923 | 18.46 |
ENSMUST00000031910.7
|
Prss1
|
protease, serine, 1 (trypsin 1) |
| chr16_-_16863975 | 16.48 |
ENSMUST00000100136.3
|
Igll1
|
immunoglobulin lambda-like polypeptide 1 |
| chr8_+_57455898 | 15.98 |
ENSMUST00000034023.3
|
Scrg1
|
scrapie responsive gene 1 |
| chr3_+_108364882 | 15.01 |
ENSMUST00000090563.5
|
Mybphl
|
myosin binding protein H-like |
| chr11_-_102365111 | 14.51 |
ENSMUST00000006749.9
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr17_+_40811089 | 12.86 |
ENSMUST00000024721.7
|
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr7_+_131032061 | 11.11 |
ENSMUST00000084509.3
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr15_+_79895017 | 10.31 |
ENSMUST00000023054.7
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr2_+_131491764 | 10.00 |
ENSMUST00000028806.5
ENSMUST00000110179.2 ENSMUST00000110189.2 ENSMUST00000110182.2 ENSMUST00000110183.2 ENSMUST00000110186.2 ENSMUST00000110188.1 |
Smox
|
spermine oxidase |
| chr19_+_58759700 | 9.51 |
ENSMUST00000026081.3
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr7_-_119459266 | 7.76 |
ENSMUST00000033255.5
|
Gp2
|
glycoprotein 2 (zymogen granule membrane) |
| chr7_-_100855403 | 7.57 |
ENSMUST00000156855.1
|
Relt
|
RELT tumor necrosis factor receptor |
| chr1_+_139454747 | 7.49 |
ENSMUST00000053364.8
ENSMUST00000097554.3 |
Aspm
|
asp (abnormal spindle)-like, microcephaly associated (Drosophila) |
| chr17_+_43568641 | 7.39 |
ENSMUST00000169694.1
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr2_+_31245801 | 7.31 |
ENSMUST00000000199.7
|
Ncs1
|
neuronal calcium sensor 1 |
| chr17_+_43568269 | 7.25 |
ENSMUST00000024706.5
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr17_+_43568475 | 7.02 |
ENSMUST00000167418.1
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr17_+_43568096 | 6.97 |
ENSMUST00000167214.1
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr5_+_122206792 | 6.88 |
ENSMUST00000145854.1
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr7_+_128062657 | 6.77 |
ENSMUST00000120355.1
ENSMUST00000106240.2 ENSMUST00000098015.3 |
Itgam
|
integrin alpha M |
| chr16_+_32756336 | 6.34 |
ENSMUST00000135753.1
|
Muc4
|
mucin 4 |
| chr3_-_75270073 | 5.88 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr2_+_127336152 | 5.57 |
ENSMUST00000028846.6
|
Dusp2
|
dual specificity phosphatase 2 |
| chr5_-_148371525 | 5.41 |
ENSMUST00000138596.1
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr7_+_128062635 | 5.39 |
ENSMUST00000064821.7
ENSMUST00000106242.3 |
Itgam
|
integrin alpha M |
| chr1_+_174158722 | 5.16 |
ENSMUST00000068403.3
|
Olfr420
|
olfactory receptor 420 |
| chrX_-_142966709 | 5.06 |
ENSMUST00000041317.2
|
Ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr6_-_40951826 | 4.85 |
ENSMUST00000073642.5
|
Gm4744
|
predicted gene 4744 |
| chr19_+_4154606 | 4.83 |
ENSMUST00000061086.8
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chrX_-_73659724 | 4.64 |
ENSMUST00000114473.1
ENSMUST00000002087.7 |
Pnck
|
pregnancy upregulated non-ubiquitously expressed CaM kinase |
| chr7_-_14123042 | 4.54 |
ENSMUST00000098809.2
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr5_+_21543525 | 4.52 |
ENSMUST00000035651.4
|
Lrrc17
|
leucine rich repeat containing 17 |
| chr18_+_62180119 | 4.51 |
ENSMUST00000067743.1
|
Gm9949
|
predicted gene 9949 |
| chr13_-_66852017 | 4.50 |
ENSMUST00000059329.6
|
Gm17449
|
predicted gene, 17449 |
| chr9_+_108991902 | 4.50 |
ENSMUST00000147989.1
ENSMUST00000051873.8 |
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr3_+_124321031 | 4.48 |
ENSMUST00000058994.4
|
Tram1l1
|
translocation associated membrane protein 1-like 1 |
| chr7_+_128062698 | 4.40 |
ENSMUST00000119696.1
|
Itgam
|
integrin alpha M |
| chr5_+_122210134 | 4.38 |
ENSMUST00000100747.2
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr13_-_66851513 | 4.27 |
ENSMUST00000169322.1
|
Gm17404
|
predicted gene, 17404 |
| chr6_+_123123313 | 4.20 |
ENSMUST00000041779.6
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr6_+_123123423 | 4.17 |
ENSMUST00000032248.7
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr7_+_81114799 | 4.08 |
ENSMUST00000026820.4
|
Slc28a1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
| chr2_+_30281043 | 3.86 |
ENSMUST00000143119.2
|
RP23-395P6.9
|
RP23-395P6.9 |
| chr5_+_77016023 | 3.85 |
ENSMUST00000031161.4
ENSMUST00000117880.1 |
1700023E05Rik
|
RIKEN cDNA 1700023E05 gene |
| chr12_-_113422730 | 3.80 |
ENSMUST00000177715.1
ENSMUST00000103426.1 |
Ighm
|
immunoglobulin heavy constant mu |
| chr11_+_98412461 | 3.71 |
ENSMUST00000058295.5
|
Erbb2
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) |
| chr10_+_128238034 | 3.67 |
ENSMUST00000105245.2
|
Timeless
|
timeless circadian clock 1 |
| chr17_+_35241746 | 3.56 |
ENSMUST00000068056.5
ENSMUST00000174757.1 |
Ddx39b
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr16_+_32735886 | 3.43 |
ENSMUST00000132475.1
ENSMUST00000096106.3 |
Muc4
|
mucin 4 |
| chr15_+_98571004 | 3.40 |
ENSMUST00000023728.6
|
4930415O20Rik
|
RIKEN cDNA 4930415O20 gene |
| chr7_-_98178254 | 3.36 |
ENSMUST00000040971.7
|
Capn5
|
calpain 5 |
| chr4_+_11758147 | 3.25 |
ENSMUST00000029871.5
ENSMUST00000108303.1 |
Cdh17
|
cadherin 17 |
| chr5_+_140607334 | 3.25 |
ENSMUST00000031555.1
|
Lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr17_+_35241838 | 3.24 |
ENSMUST00000173731.1
|
Ddx39b
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr3_+_87376381 | 3.23 |
ENSMUST00000163661.1
ENSMUST00000072480.2 ENSMUST00000167200.1 |
Fcrl1
|
Fc receptor-like 1 |
| chr6_+_120093348 | 3.15 |
ENSMUST00000112711.2
|
Ninj2
|
ninjurin 2 |
| chr13_-_49248146 | 3.14 |
ENSMUST00000119721.1
ENSMUST00000058196.6 |
Susd3
|
sushi domain containing 3 |
| chr7_+_43437073 | 2.96 |
ENSMUST00000070518.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr19_-_11660516 | 2.93 |
ENSMUST00000135994.1
ENSMUST00000121793.1 ENSMUST00000069681.3 |
Plac1l
|
placenta-specific 1-like |
| chr9_+_106222598 | 2.91 |
ENSMUST00000062241.9
|
Tlr9
|
toll-like receptor 9 |
| chr8_+_69832633 | 2.77 |
ENSMUST00000131637.2
ENSMUST00000081503.6 |
Pbx4
|
pre B cell leukemia homeobox 4 |
| chr7_+_46847128 | 2.76 |
ENSMUST00000005051.4
|
Ldha
|
lactate dehydrogenase A |
| chr13_-_56178864 | 2.72 |
ENSMUST00000169652.1
|
Tifab
|
TRAF-interacting protein with forkhead-associated domain, family member B |
| chr4_-_42853888 | 2.68 |
ENSMUST00000107979.1
|
Gm12429
|
predicted gene 12429 |
| chr2_+_106693185 | 2.60 |
ENSMUST00000111063.1
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr8_+_21734490 | 2.58 |
ENSMUST00000080533.5
|
Defa24
|
defensin, alpha, 24 |
| chr3_+_92316705 | 2.50 |
ENSMUST00000061038.2
|
Sprr2b
|
small proline-rich protein 2B |
| chr1_-_165708088 | 2.50 |
ENSMUST00000040357.8
ENSMUST00000097474.3 |
Rcsd1
|
RCSD domain containing 1 |
| chr6_-_50382831 | 2.49 |
ENSMUST00000114468.2
|
Osbpl3
|
oxysterol binding protein-like 3 |
| chr16_+_23290464 | 2.45 |
ENSMUST00000115335.1
|
St6gal1
|
beta galactoside alpha 2,6 sialyltransferase 1 |
| chr18_+_35553401 | 2.45 |
ENSMUST00000181664.1
|
Snhg4
|
small nucleolar RNA host gene 4 (non-protein coding) |
| chr10_-_86732409 | 2.44 |
ENSMUST00000070435.4
|
Fabp3-ps1
|
fatty acid binding protein 3, muscle and heart, pseudogene 1 |
| chr11_+_106276715 | 2.42 |
ENSMUST00000044462.3
|
Tcam1
|
testicular cell adhesion molecule 1 |
| chr6_+_48537560 | 2.36 |
ENSMUST00000040361.5
|
Atp6v0e2
|
ATPase, H+ transporting, lysosomal V0 subunit E2 |
| chr5_-_108795352 | 2.34 |
ENSMUST00000004943.1
|
Tmed11
|
transmembrane emp24 protein transport domain containing |
| chr17_+_57279094 | 2.34 |
ENSMUST00000169220.2
ENSMUST00000005889.9 ENSMUST00000112870.4 |
Vav1
|
vav 1 oncogene |
| chr3_-_37724321 | 2.33 |
ENSMUST00000108105.1
ENSMUST00000079755.4 ENSMUST00000099128.1 |
Gm5148
|
predicted gene 5148 |
| chr7_+_19577287 | 2.31 |
ENSMUST00000108453.1
|
Zfp296
|
zinc finger protein 296 |
| chr15_-_74763567 | 2.27 |
ENSMUST00000040404.6
|
Ly6d
|
lymphocyte antigen 6 complex, locus D |
| chr15_-_82244716 | 2.16 |
ENSMUST00000089155.4
ENSMUST00000089157.3 |
Cenpm
|
centromere protein M |
| chr12_+_105705970 | 2.15 |
ENSMUST00000040876.5
|
Ak7
|
adenylate kinase 7 |
| chr16_-_90810365 | 2.13 |
ENSMUST00000140920.1
|
Urb1
|
URB1 ribosome biogenesis 1 homolog (S. cerevisiae) |
| chr1_-_65123108 | 2.09 |
ENSMUST00000050047.3
ENSMUST00000148020.1 |
D630023F18Rik
|
RIKEN cDNA D630023F18 gene |
| chr3_-_101287879 | 2.09 |
ENSMUST00000152321.1
|
Cd2
|
CD2 antigen |
| chr5_+_25246775 | 2.09 |
ENSMUST00000144971.1
|
Galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 |
| chr6_-_52218686 | 2.09 |
ENSMUST00000134367.2
|
Hoxa7
|
homeobox A7 |
| chr11_-_6230127 | 2.05 |
ENSMUST00000004505.2
|
Npc1l1
|
NPC1-like 1 |
| chr7_+_43950614 | 2.03 |
ENSMUST00000072204.4
|
Klk1b8
|
kallikrein 1-related peptidase b8 |
| chr3_-_101287897 | 2.00 |
ENSMUST00000029456.4
|
Cd2
|
CD2 antigen |
| chr11_-_8973266 | 1.95 |
ENSMUST00000154153.1
|
Pkd1l1
|
polycystic kidney disease 1 like 1 |
| chr15_-_63902936 | 1.92 |
ENSMUST00000063530.6
|
Gsdmc4
|
gasdermin C4 |
| chr19_+_11407652 | 1.91 |
ENSMUST00000072729.3
|
Ms4a4c
|
membrane-spanning 4-domains, subfamily A, member 4C |
| chr3_+_130068390 | 1.91 |
ENSMUST00000076703.6
|
Gm9396
|
predicted gene 9396 |
| chrX_+_159627265 | 1.90 |
ENSMUST00000112456.2
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr19_-_32196393 | 1.83 |
ENSMUST00000151822.1
|
Sgms1
|
sphingomyelin synthase 1 |
| chr10_-_81627797 | 1.83 |
ENSMUST00000042923.8
|
Sirt6
|
sirtuin 6 |
| chr13_+_112800878 | 1.77 |
ENSMUST00000016144.4
|
Ppap2a
|
phosphatidic acid phosphatase type 2A |
| chr1_+_165485168 | 1.68 |
ENSMUST00000111440.1
ENSMUST00000027852.8 ENSMUST00000111439.1 |
Adcy10
|
adenylate cyclase 10 |
| chr7_+_66839726 | 1.67 |
ENSMUST00000098382.3
|
Adamts17
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 17 |
| chr17_+_17316078 | 1.67 |
ENSMUST00000105311.3
|
Gm6712
|
predicted gene 6712 |
| chr9_-_119341390 | 1.63 |
ENSMUST00000139870.1
|
Myd88
|
myeloid differentiation primary response gene 88 |
| chr15_+_82252397 | 1.61 |
ENSMUST00000136948.1
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr1_-_128359610 | 1.58 |
ENSMUST00000027601.4
|
Mcm6
|
minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) |
| chr14_+_58893465 | 1.50 |
ENSMUST00000079960.1
|
Rpl13-ps3
|
ribosomal protein L13, pseudogene 3 |
| chr1_-_165634451 | 1.50 |
ENSMUST00000111435.2
ENSMUST00000068705.7 |
Mpzl1
|
myelin protein zero-like 1 |
| chrX_-_74373260 | 1.49 |
ENSMUST00000073067.4
ENSMUST00000037967.5 |
Slc10a3
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 3 |
| chr11_+_115564434 | 1.48 |
ENSMUST00000021085.4
|
Nup85
|
nucleoporin 85 |
| chr9_-_114564315 | 1.47 |
ENSMUST00000111816.2
|
Trim71
|
tripartite motif-containing 71 |
| chr2_-_114175321 | 1.42 |
ENSMUST00000043160.6
|
Aqr
|
aquarius |
| chr5_+_121204477 | 1.41 |
ENSMUST00000031617.9
|
Rpl6
|
ribosomal protein L6 |
| chr11_-_69837781 | 1.38 |
ENSMUST00000108634.2
|
Nlgn2
|
neuroligin 2 |
| chr13_+_94083490 | 1.37 |
ENSMUST00000156071.1
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chr11_-_46166397 | 1.36 |
ENSMUST00000020679.2
|
Nipal4
|
NIPA-like domain containing 4 |
| chr4_+_129906205 | 1.35 |
ENSMUST00000145196.1
ENSMUST00000141731.1 ENSMUST00000129149.1 |
E330017L17Rik
|
RIKEN cDNA E330017L17 gene |
| chr1_-_23922283 | 1.35 |
ENSMUST00000027339.7
|
Smap1
|
small ArfGAP 1 |
| chr4_+_5644084 | 1.35 |
ENSMUST00000054857.6
|
Fam110b
|
family with sequence similarity 110, member B |
| chr8_-_106573461 | 1.33 |
ENSMUST00000073722.5
|
Gm10073
|
predicted pseudogene 10073 |
| chr8_+_69822429 | 1.23 |
ENSMUST00000164890.1
ENSMUST00000034325.4 |
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr6_-_57844493 | 1.22 |
ENSMUST00000081186.3
|
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr12_-_87443800 | 1.19 |
ENSMUST00000162961.1
|
Alkbh1
|
alkB, alkylation repair homolog 1 (E. coli) |
| chrX_-_7947763 | 1.18 |
ENSMUST00000154244.1
|
Hdac6
|
histone deacetylase 6 |
| chr5_+_139211934 | 1.17 |
ENSMUST00000148772.1
ENSMUST00000110882.1 |
Sun1
|
Sad1 and UNC84 domain containing 1 |
| chr18_-_88927447 | 1.16 |
ENSMUST00000147313.1
|
Socs6
|
suppressor of cytokine signaling 6 |
| chr4_-_120951664 | 1.14 |
ENSMUST00000106280.1
|
Zfp69
|
zinc finger protein 69 |
| chr13_+_100124850 | 1.12 |
ENSMUST00000022147.8
ENSMUST00000091321.5 ENSMUST00000143937.1 |
Smn1
|
survival motor neuron 1 |
| chr19_-_53589067 | 1.10 |
ENSMUST00000095978.3
|
Nutf2-ps1
|
nuclear transport factor 2, pseudogene 1 |
| chr7_-_101903783 | 1.10 |
ENSMUST00000106969.1
|
Tomt
|
transmembrane O-methyltransferase |
| chr18_+_74216118 | 1.07 |
ENSMUST00000025444.6
|
Cxxc1
|
CXXC finger 1 (PHD domain) |
| chr2_-_110950923 | 1.05 |
ENSMUST00000099623.3
|
Ano3
|
anoctamin 3 |
| chr7_-_42578588 | 1.03 |
ENSMUST00000179470.1
|
Gm21028
|
predicted gene, 21028 |
| chr8_+_40423786 | 1.01 |
ENSMUST00000049389.4
ENSMUST00000128166.1 ENSMUST00000167766.1 |
Zdhhc2
|
zinc finger, DHHC domain containing 2 |
| chr19_-_11336107 | 1.01 |
ENSMUST00000056035.2
ENSMUST00000067532.4 |
Ms4a7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr2_-_114175274 | 1.00 |
ENSMUST00000102543.4
|
Aqr
|
aquarius |
| chr1_+_171682004 | 1.00 |
ENSMUST00000015499.7
ENSMUST00000068584.5 |
Cd48
|
CD48 antigen |
| chr16_-_19515634 | 0.98 |
ENSMUST00000054606.1
|
Olfr167
|
olfactory receptor 167 |
| chr16_+_11203375 | 0.97 |
ENSMUST00000181526.1
|
2610020C07Rik
|
RIKEN cDNA 2610020C07 gene |
| chrX_-_7947553 | 0.95 |
ENSMUST00000133349.1
|
Hdac6
|
histone deacetylase 6 |
| chr10_-_97726755 | 0.93 |
ENSMUST00000166373.1
|
C030005K15Rik
|
RIKEN cDNA C030005K15 gene |
| chr4_+_42318334 | 0.93 |
ENSMUST00000178192.1
|
Gm21598
|
predicted gene, 21598 |
| chr11_+_60353324 | 0.91 |
ENSMUST00000070805.6
ENSMUST00000094140.2 ENSMUST00000108723.2 ENSMUST00000108722.4 |
Lrrc48
|
leucine rich repeat containing 48 |
| chr8_+_85026833 | 0.88 |
ENSMUST00000047281.8
|
2310036O22Rik
|
RIKEN cDNA 2310036O22 gene |
| chrX_-_105929397 | 0.82 |
ENSMUST00000113573.1
ENSMUST00000130980.1 |
Atrx
|
alpha thalassemia/mental retardation syndrome X-linked homolog (human) |
| chr1_+_40266578 | 0.82 |
ENSMUST00000114795.1
|
Il1r1
|
interleukin 1 receptor, type I |
| chr3_-_95133989 | 0.81 |
ENSMUST00000172572.2
ENSMUST00000173462.2 |
Scnm1
|
sodium channel modifier 1 |
| chr11_+_101987050 | 0.78 |
ENSMUST00000010985.7
|
1700006E09Rik
|
RIKEN cDNA 1700006E09 gene |
| chr4_-_88676924 | 0.78 |
ENSMUST00000105148.1
|
Gm13280
|
predicted gene 13280 |
| chr3_-_158016419 | 0.78 |
ENSMUST00000127778.1
|
Srsf11
|
serine/arginine-rich splicing factor 11 |
| chr19_+_25672408 | 0.78 |
ENSMUST00000053068.5
|
Dmrt2
|
doublesex and mab-3 related transcription factor 2 |
| chr11_-_87826023 | 0.77 |
ENSMUST00000103177.3
|
Lpo
|
lactoperoxidase |
| chr16_+_10835046 | 0.76 |
ENSMUST00000037913.8
|
Rmi2
|
RMI2, RecQ mediated genome instability 2, homolog (S. cerevisiae) |
| chr2_-_127482499 | 0.73 |
ENSMUST00000088538.5
|
Kcnip3
|
Kv channel interacting protein 3, calsenilin |
| chr18_+_32815383 | 0.72 |
ENSMUST00000025237.3
|
Tslp
|
thymic stromal lymphopoietin |
| chr2_-_13271268 | 0.71 |
ENSMUST00000137670.1
ENSMUST00000114791.2 |
Rsu1
|
Ras suppressor protein 1 |
| chr8_-_94918012 | 0.69 |
ENSMUST00000077955.5
|
Ccdc102a
|
coiled-coil domain containing 102A |
| chr6_+_57002300 | 0.68 |
ENSMUST00000079669.4
|
Vmn1r6
|
vomeronasal 1 receptor 6 |
| chr6_+_129397297 | 0.66 |
ENSMUST00000032262.7
|
Clec1b
|
C-type lectin domain family 1, member b |
| chr1_-_118982551 | 0.64 |
ENSMUST00000159678.1
|
Gli2
|
GLI-Kruppel family member GLI2 |
| chr15_-_63869297 | 0.62 |
ENSMUST00000089894.5
|
Gsdmc3
|
gasdermin C3 |
| chr12_+_87954475 | 0.62 |
ENSMUST00000181843.1
ENSMUST00000180706.1 ENSMUST00000181394.1 ENSMUST00000110145.4 ENSMUST00000181326.1 ENSMUST00000181300.1 |
Gm2042
|
predicted gene 2042 |
| chr10_+_24595623 | 0.60 |
ENSMUST00000176228.1
ENSMUST00000129142.1 |
Ctgf
|
connective tissue growth factor |
| chr4_+_102986343 | 0.59 |
ENSMUST00000030248.5
ENSMUST00000125417.1 |
Tctex1d1
|
Tctex1 domain containing 1 |
| chrX_-_7947848 | 0.59 |
ENSMUST00000115642.1
ENSMUST00000033501.8 ENSMUST00000145675.1 |
Hdac6
|
histone deacetylase 6 |
| chr2_-_13271419 | 0.58 |
ENSMUST00000028059.2
|
Rsu1
|
Ras suppressor protein 1 |
| chr12_-_103989917 | 0.58 |
ENSMUST00000151709.2
ENSMUST00000176246.1 ENSMUST00000074693.7 |
Serpina11
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
| chr15_+_55557575 | 0.56 |
ENSMUST00000170046.1
|
Mtbp
|
Mdm2, transformed 3T3 cell double minute p53 binding protein |
| chr19_-_4625612 | 0.54 |
ENSMUST00000025823.3
|
Rce1
|
RCE1 homolog, prenyl protein peptidase (S. cerevisiae) |
| chr5_+_117319258 | 0.53 |
ENSMUST00000111967.1
|
Vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr11_-_83592981 | 0.52 |
ENSMUST00000019071.3
|
Ccl6
|
chemokine (C-C motif) ligand 6 |
| chr14_+_55510445 | 0.50 |
ENSMUST00000165262.1
ENSMUST00000074225.4 |
Cpne6
|
copine VI |
| chr7_+_82611777 | 0.49 |
ENSMUST00000172784.1
|
Adamtsl3
|
ADAMTS-like 3 |
| chr7_-_118243564 | 0.49 |
ENSMUST00000179047.1
ENSMUST00000032891.8 |
Smg1
|
SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) |
| chr2_-_119029359 | 0.48 |
ENSMUST00000110833.1
|
Ccdc32
|
coiled-coil domain containing 32 |
| chrX_-_74373218 | 0.48 |
ENSMUST00000178691.1
ENSMUST00000114146.1 |
Ubl4
Slc10a3
|
ubiquitin-like 4 solute carrier family 10 (sodium/bile acid cotransporter family), member 3 |
| chr9_+_98422961 | 0.47 |
ENSMUST00000052068.9
|
Rbp1
|
retinol binding protein 1, cellular |
| chr8_+_21134610 | 0.44 |
ENSMUST00000098898.4
|
Gm15284
|
predicted gene 15284 |
| chr4_+_156013835 | 0.40 |
ENSMUST00000030952.5
|
Tnfrsf4
|
tumor necrosis factor receptor superfamily, member 4 |
| chr1_+_60746358 | 0.32 |
ENSMUST00000027165.2
|
Cd28
|
CD28 antigen |
| chrX_-_12673540 | 0.32 |
ENSMUST00000060108.6
|
1810030O07Rik
|
RIKEN cDNA 1810030O07 gene |
| chr7_+_123123870 | 0.31 |
ENSMUST00000094053.5
|
Tnrc6a
|
trinucleotide repeat containing 6a |
| chr14_+_70555900 | 0.30 |
ENSMUST00000163060.1
|
Hr
|
hairless |
| chr12_-_103989950 | 0.27 |
ENSMUST00000120251.2
|
Serpina11
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 |
| chr8_+_31150307 | 0.26 |
ENSMUST00000098842.2
|
Tti2
|
TELO2 interacting protein 2 |
| chr4_+_150148905 | 0.24 |
ENSMUST00000059893.7
|
Slc2a7
|
solute carrier family 2 (facilitated glucose transporter), member 7 |
| chr8_-_86885222 | 0.19 |
ENSMUST00000034074.7
|
N4bp1
|
NEDD4 binding protein 1 |
| chr2_+_86074715 | 0.19 |
ENSMUST00000164985.1
|
Olfr1036
|
olfactory receptor 1036 |
| chr11_-_4849345 | 0.19 |
ENSMUST00000053079.6
ENSMUST00000109910.2 |
Nf2
|
neurofibromatosis 2 |
| chr1_+_60909148 | 0.19 |
ENSMUST00000097720.3
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr5_-_129879038 | 0.19 |
ENSMUST00000026617.6
|
Phkg1
|
phosphorylase kinase gamma 1 |
| chr17_-_62606679 | 0.18 |
ENSMUST00000163332.1
|
Efna5
|
ephrin A5 |
| chrX_+_169685191 | 0.16 |
ENSMUST00000112104.1
ENSMUST00000112107.1 |
Mid1
|
midline 1 |
| chr19_+_8764934 | 0.16 |
ENSMUST00000184663.1
|
Nxf1
|
nuclear RNA export factor 1 |
| chr5_-_23616528 | 0.15 |
ENSMUST00000088392.4
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr11_-_99230998 | 0.14 |
ENSMUST00000103133.3
|
Smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr5_+_140419248 | 0.12 |
ENSMUST00000100507.3
|
Eif3b
|
eukaryotic translation initiation factor 3, subunit B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Six6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.4 | 37.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 4.8 | 28.6 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 3.0 | 20.8 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 2.4 | 16.6 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 1.6 | 9.8 | GO:0002835 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) |
| 1.6 | 7.8 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 1.5 | 7.5 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 1.5 | 2.9 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 1.4 | 10.0 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 1.3 | 3.8 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 1.2 | 3.7 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 1.1 | 3.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 1.0 | 11.3 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 1.0 | 4.1 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 1.0 | 6.8 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.9 | 15.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.9 | 2.7 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.7 | 3.7 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.7 | 10.3 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.7 | 2.7 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 0.7 | 2.0 | GO:0009726 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.6 | 3.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.6 | 2.5 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.6 | 1.8 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.6 | 11.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.6 | 4.5 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.6 | 9.5 | GO:0019377 | galactolipid metabolic process(GO:0019374) glycolipid catabolic process(GO:0019377) |
| 0.5 | 14.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.5 | 4.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.5 | 5.4 | GO:0015809 | arginine transport(GO:0015809) |
| 0.5 | 16.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.5 | 2.8 | GO:0019661 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.4 | 59.6 | GO:0007586 | digestion(GO:0007586) |
| 0.4 | 1.6 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.3 | 4.1 | GO:2000484 | positive regulation of interleukin-8 secretion(GO:2000484) |
| 0.3 | 2.5 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 0.3 | 1.2 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.3 | 1.4 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.3 | 2.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.2 | 0.8 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.2 | 1.8 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 0.8 | GO:0061055 | myotome development(GO:0061055) |
| 0.2 | 2.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.2 | 0.6 | GO:0060032 | notochord regression(GO:0060032) |
| 0.2 | 1.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 0.5 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.2 | 1.1 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 | 1.1 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 | 0.8 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 5.6 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.1 | 0.5 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.6 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 | 1.8 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 17.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 2.3 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 4.0 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.1 | 2.8 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.1 | 1.6 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 1.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 2.5 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.1 | 7.3 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.1 | 2.3 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.1 | 6.7 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 1.3 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 1.9 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.4 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.2 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 1.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0035128 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.0 | 1.5 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 4.8 | GO:0018210 | peptidyl-threonine modification(GO:0018210) |
| 0.0 | 0.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:1903978 | regulation of microglial cell activation(GO:1903978) |
| 0.0 | 0.7 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 6.3 | GO:0032496 | response to lipopolysaccharide(GO:0032496) |
| 0.0 | 0.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.8 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 1.1 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.7 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 2.0 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 1.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.2 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.0 | 0.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.5 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 1.2 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.0 | 1.7 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.5 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.0 | GO:0002819 | regulation of adaptive immune response(GO:0002819) |
| 0.0 | 0.8 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 1.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 11.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 1.5 | 7.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 1.3 | 28.6 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 1.1 | 20.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 1.0 | 2.9 | GO:0036019 | endolysosome(GO:0036019) |
| 0.7 | 9.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.6 | 6.8 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.6 | 15.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.4 | 4.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.4 | 9.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.4 | 7.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.4 | 16.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.3 | 3.7 | GO:0043219 | lateral loop(GO:0043219) |
| 0.3 | 2.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.2 | 1.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.2 | 2.4 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 14.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 2.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.5 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 1.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 2.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 2.8 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 5.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 1.7 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 10.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.8 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 16.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 2.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.6 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 2.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 2.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 10.0 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 1.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.4 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 3.1 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.3 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 2.0 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.5 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 34.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 2.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 32.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.6 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 1.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 30.8 | GO:0005576 | extracellular region(GO:0005576) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 10.0 | GO:0052901 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 2.9 | 28.6 | GO:0047499 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) calcium-independent phospholipase A2 activity(GO:0047499) |
| 2.6 | 10.3 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 2.4 | 9.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 2.4 | 16.6 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 2.3 | 20.8 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 2.3 | 11.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 1.4 | 4.1 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 1.4 | 6.8 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 1.3 | 37.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 1.2 | 3.7 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 1.1 | 3.2 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 1.1 | 3.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.8 | 20.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.8 | 15.0 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.7 | 14.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.7 | 2.7 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.7 | 5.4 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.6 | 4.5 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.6 | 9.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.6 | 1.8 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.6 | 4.0 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.5 | 2.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.5 | 4.5 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.5 | 2.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.4 | 1.6 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.4 | 2.8 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.4 | 5.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.4 | 1.5 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.4 | 1.8 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.4 | 1.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.3 | 2.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 0.8 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 8.0 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 1.2 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.2 | 1.4 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 19.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 37.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 8.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 12.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 7.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 3.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 4.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 2.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 2.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 1.9 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.1 | 2.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 1.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 6.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.5 | GO:0016918 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.0 | 1.1 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 17.5 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 6.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 4.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 2.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.8 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 2.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.3 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 0.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 28.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.5 | 16.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 3.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 3.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 27.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 7.3 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 2.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 10.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 10.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 4.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 24.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.6 | 18.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.6 | 10.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.5 | 19.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.4 | 9.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.4 | 12.9 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.3 | 4.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.3 | 2.7 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 5.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 21.0 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.2 | 14.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 9.5 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.2 | 3.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 2.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 3.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 1.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 4.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 3.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 2.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 2.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 2.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.9 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 2.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |