Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Ptf1a
Z-value: 1.45
Transcription factors associated with Ptf1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ptf1a
|
ENSMUSG00000026735.2 | pancreas specific transcription factor, 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ptf1a | mm10_v2_chr2_+_19445632_19445663 | 0.78 | 2.7e-08 | Click! |
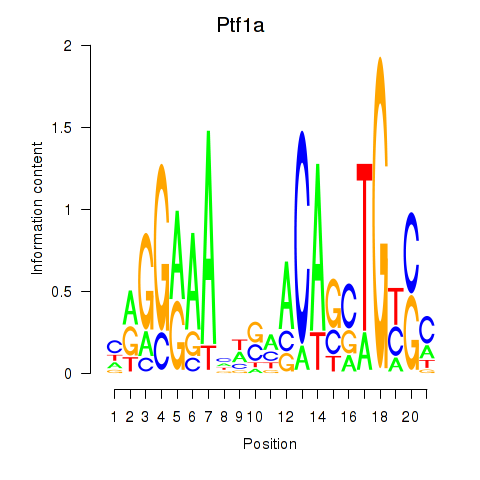
Activity profile of Ptf1a motif
Sorted Z-values of Ptf1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_41314700 | 29.30 |
ENSMUST00000064324.5
|
Try5
|
trypsin 5 |
| chr6_+_41458923 | 22.61 |
ENSMUST00000031910.7
|
Prss1
|
protease, serine, 1 (trypsin 1) |
| chr6_+_41392356 | 22.34 |
ENSMUST00000049079.7
|
Gm5771
|
predicted gene 5771 |
| chr7_+_28540863 | 18.76 |
ENSMUST00000119180.2
|
Sycn
|
syncollin |
| chr6_+_41354105 | 18.51 |
ENSMUST00000072103.5
|
Try10
|
trypsin 10 |
| chr6_+_41302265 | 16.94 |
ENSMUST00000031913.4
|
Try4
|
trypsin 4 |
| chr7_-_131322292 | 15.52 |
ENSMUST00000046611.7
|
Cuzd1
|
CUB and zona pellucida-like domains 1 |
| chr2_+_173153048 | 7.25 |
ENSMUST00000029017.5
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr6_-_69243445 | 6.43 |
ENSMUST00000101325.3
|
Igkv4-71
|
immunoglobulin kappa chain variable 4-71 |
| chr1_-_90153396 | 3.53 |
ENSMUST00000113094.2
|
Iqca
|
IQ motif containing with AAA domain |
| chr10_+_95417352 | 3.53 |
ENSMUST00000181781.1
|
5730420D15Rik
|
RIKEN cDNA 5730420D15 gene |
| chr14_-_63417125 | 3.36 |
ENSMUST00000014597.3
|
Blk
|
B lymphoid kinase |
| chr11_+_101330605 | 3.03 |
ENSMUST00000103105.3
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr8_+_105048592 | 2.91 |
ENSMUST00000093222.5
ENSMUST00000093223.3 |
Ces3a
|
carboxylesterase 3A |
| chrX_+_160390684 | 2.64 |
ENSMUST00000112408.2
ENSMUST00000112402.1 ENSMUST00000112401.1 ENSMUST00000112400.1 ENSMUST00000112405.2 ENSMUST00000112404.2 ENSMUST00000146805.1 |
Gpr64
|
G protein-coupled receptor 64 |
| chrX_-_162964557 | 2.07 |
ENSMUST00000038769.2
|
S100g
|
S100 calcium binding protein G |
| chr7_-_3249711 | 1.92 |
ENSMUST00000108653.2
|
Nlrp12
|
NLR family, pyrin domain containing 12 |
| chr15_-_77399086 | 1.74 |
ENSMUST00000175919.1
ENSMUST00000176074.1 |
Apol7a
|
apolipoprotein L 7a |
| chr14_-_33185489 | 1.72 |
ENSMUST00000159606.1
|
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr11_+_120633719 | 1.67 |
ENSMUST00000181502.1
|
Gm17586
|
predicted gene, 17586 |
| chr3_+_138277489 | 1.54 |
ENSMUST00000004232.9
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr11_+_4031770 | 1.51 |
ENSMUST00000019512.7
|
Sec14l4
|
SEC14-like 4 (S. cerevisiae) |
| chr11_-_99155067 | 1.40 |
ENSMUST00000103134.3
|
Ccr7
|
chemokine (C-C motif) receptor 7 |
| chr10_-_95417099 | 1.36 |
ENSMUST00000135822.1
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr2_+_164948219 | 1.35 |
ENSMUST00000017881.2
|
Mmp9
|
matrix metallopeptidase 9 |
| chr16_+_91269759 | 1.24 |
ENSMUST00000056882.5
|
Olig1
|
oligodendrocyte transcription factor 1 |
| chr7_-_30944017 | 1.20 |
ENSMUST00000062620.7
|
Hamp
|
hepcidin antimicrobial peptide |
| chr7_-_140102367 | 1.14 |
ENSMUST00000142105.1
|
Fuom
|
fucose mutarotase |
| chr2_+_11705712 | 1.03 |
ENSMUST00000138856.1
ENSMUST00000078834.5 ENSMUST00000114834.3 ENSMUST00000114833.3 ENSMUST00000114831.2 ENSMUST00000114832.2 |
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr10_-_77089428 | 1.01 |
ENSMUST00000156009.1
|
Col18a1
|
collagen, type XVIII, alpha 1 |
| chr2_-_148038270 | 1.00 |
ENSMUST00000132070.1
|
9030622O22Rik
|
RIKEN cDNA 9030622O22 gene |
| chr14_-_55944536 | 0.97 |
ENSMUST00000022834.6
|
Cma1
|
chymase 1, mast cell |
| chr11_-_90390895 | 0.92 |
ENSMUST00000004051.7
|
Hlf
|
hepatic leukemia factor |
| chr11_+_78465697 | 0.87 |
ENSMUST00000001126.3
|
Slc46a1
|
solute carrier family 46, member 1 |
| chr14_+_52792208 | 0.85 |
ENSMUST00000178426.2
|
Trav9d-1
|
T cell receptor alpha variable 9D-1 |
| chr17_-_28517509 | 0.85 |
ENSMUST00000114792.1
ENSMUST00000177939.1 |
Fkbp5
|
FK506 binding protein 5 |
| chr11_-_6444352 | 0.84 |
ENSMUST00000093346.5
ENSMUST00000109737.2 |
H2afv
|
H2A histone family, member V |
| chr14_-_70207637 | 0.84 |
ENSMUST00000022682.5
|
Sorbs3
|
sorbin and SH3 domain containing 3 |
| chr6_-_136781718 | 0.83 |
ENSMUST00000078095.6
ENSMUST00000032338.7 |
Gucy2c
|
guanylate cyclase 2c |
| chr10_+_127048235 | 0.82 |
ENSMUST00000165764.1
|
Cyp27b1
|
cytochrome P450, family 27, subfamily b, polypeptide 1 |
| chr5_-_120887582 | 0.82 |
ENSMUST00000086368.5
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G |
| chr5_-_99252839 | 0.77 |
ENSMUST00000168092.1
ENSMUST00000031276.8 |
Rasgef1b
|
RasGEF domain family, member 1B |
| chr2_-_30415302 | 0.77 |
ENSMUST00000132981.2
ENSMUST00000129494.1 |
Crat
|
carnitine acetyltransferase |
| chr10_-_75932468 | 0.72 |
ENSMUST00000120281.1
ENSMUST00000000924.6 |
Mmp11
|
matrix metallopeptidase 11 |
| chr9_-_106476590 | 0.68 |
ENSMUST00000112479.2
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr4_-_155010984 | 0.66 |
ENSMUST00000105631.2
ENSMUST00000139976.2 ENSMUST00000145662.2 |
Plch2
|
phospholipase C, eta 2 |
| chr7_+_104329471 | 0.63 |
ENSMUST00000180136.1
ENSMUST00000178316.1 |
Trim34b
|
tripartite motif-containing 34B |
| chr15_+_31276491 | 0.61 |
ENSMUST00000068987.5
|
Fam136b-ps
|
family with sequence similarity 136, member B, pseudogene |
| chr9_-_108597558 | 0.59 |
ENSMUST00000006853.5
|
P4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr19_+_7056731 | 0.56 |
ENSMUST00000040261.5
|
Macrod1
|
MACRO domain containing 1 |
| chr7_+_118600152 | 0.55 |
ENSMUST00000121744.1
|
Tmc5
|
transmembrane channel-like gene family 5 |
| chr11_+_66957894 | 0.54 |
ENSMUST00000127166.1
|
9130409J20Rik
|
RIKEN cDNA 9130409J20 gene |
| chr13_-_60864373 | 0.54 |
ENSMUST00000091569.5
|
4930486L24Rik
|
RIKEN cDNA 4930486L24 gene |
| chr5_-_31048014 | 0.53 |
ENSMUST00000137223.1
|
Slc5a6
|
solute carrier family 5 (sodium-dependent vitamin transporter), member 6 |
| chr7_+_28756138 | 0.52 |
ENSMUST00000178767.1
|
Gm6537
|
predicted gene 6537 |
| chr15_-_99087817 | 0.52 |
ENSMUST00000064462.3
|
C1ql4
|
complement component 1, q subcomponent-like 4 |
| chr11_+_82911253 | 0.52 |
ENSMUST00000164945.1
ENSMUST00000018989.7 |
Unc45b
|
unc-45 homolog B (C. elegans) |
| chr7_-_13054514 | 0.51 |
ENSMUST00000182087.1
|
Mzf1
|
myeloid zinc finger 1 |
| chr8_+_22283441 | 0.50 |
ENSMUST00000077194.1
|
Tpte
|
transmembrane phosphatase with tensin homology |
| chr4_+_155839675 | 0.47 |
ENSMUST00000141883.1
|
Mxra8
|
matrix-remodelling associated 8 |
| chr4_-_64046925 | 0.46 |
ENSMUST00000107377.3
|
Tnc
|
tenascin C |
| chr1_+_20951666 | 0.43 |
ENSMUST00000038447.4
|
Efhc1
|
EF-hand domain (C-terminal) containing 1 |
| chr11_-_118355496 | 0.42 |
ENSMUST00000017610.3
|
Timp2
|
tissue inhibitor of metalloproteinase 2 |
| chr14_-_33185066 | 0.42 |
ENSMUST00000061753.8
ENSMUST00000130509.2 |
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr9_+_123021315 | 0.42 |
ENSMUST00000084733.5
|
Tmem42
|
transmembrane protein 42 |
| chr17_-_27513341 | 0.42 |
ENSMUST00000118161.1
|
Grm4
|
glutamate receptor, metabotropic 4 |
| chr9_-_108305941 | 0.41 |
ENSMUST00000044725.7
|
Tcta
|
T cell leukemia translocation altered gene |
| chr8_-_105295934 | 0.40 |
ENSMUST00000057855.3
|
Exoc3l
|
exocyst complex component 3-like |
| chr11_-_96916448 | 0.38 |
ENSMUST00000103152.4
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr10_+_112083345 | 0.38 |
ENSMUST00000148897.1
ENSMUST00000020434.3 |
Glipr1l2
|
GLI pathogenesis-related 1 like 2 |
| chr16_-_44332925 | 0.36 |
ENSMUST00000136381.1
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr11_-_103344651 | 0.36 |
ENSMUST00000041385.7
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr3_-_141931523 | 0.34 |
ENSMUST00000106232.1
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr11_+_118443471 | 0.33 |
ENSMUST00000133558.1
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr11_-_59079710 | 0.32 |
ENSMUST00000133040.1
|
Obscn
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr9_-_44735189 | 0.31 |
ENSMUST00000034611.8
|
Phldb1
|
pleckstrin homology-like domain, family B, member 1 |
| chr5_+_134099704 | 0.30 |
ENSMUST00000016088.8
|
Gatsl2
|
GATS protein-like 2 |
| chr8_-_120177440 | 0.28 |
ENSMUST00000048786.6
|
Fam92b
|
family with sequence similarity 92, member B |
| chr15_-_76616841 | 0.27 |
ENSMUST00000073428.5
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr8_-_79248537 | 0.27 |
ENSMUST00000034109.4
|
1700011L22Rik
|
RIKEN cDNA 1700011L22 gene |
| chr12_+_78691516 | 0.26 |
ENSMUST00000077968.4
|
Fam71d
|
family with sequence similarity 71, member D |
| chr11_-_96916407 | 0.26 |
ENSMUST00000130774.1
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr7_-_99182681 | 0.26 |
ENSMUST00000033001.4
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr2_+_71981184 | 0.24 |
ENSMUST00000090826.5
ENSMUST00000102698.3 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr8_+_70863127 | 0.22 |
ENSMUST00000050921.2
|
A230052G05Rik
|
RIKEN cDNA A230052G05 gene |
| chr7_-_25718976 | 0.22 |
ENSMUST00000002683.2
|
Ccdc97
|
coiled-coil domain containing 97 |
| chr6_-_5256226 | 0.21 |
ENSMUST00000125686.1
ENSMUST00000031773.2 |
Pon3
|
paraoxonase 3 |
| chr4_+_152297205 | 0.21 |
ENSMUST00000048892.7
|
Icmt
|
isoprenylcysteine carboxyl methyltransferase |
| chr11_+_49247462 | 0.20 |
ENSMUST00000109194.1
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chrX_+_101383726 | 0.19 |
ENSMUST00000119190.1
|
Gjb1
|
gap junction protein, beta 1 |
| chr11_-_96916366 | 0.19 |
ENSMUST00000144731.1
ENSMUST00000127048.1 |
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr17_+_47769191 | 0.17 |
ENSMUST00000160373.1
ENSMUST00000159641.1 |
Tfeb
|
transcription factor EB |
| chr15_-_79774408 | 0.15 |
ENSMUST00000023055.6
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr18_+_37477768 | 0.15 |
ENSMUST00000051442.5
|
Pcdhb16
|
protocadherin beta 16 |
| chr9_+_106477269 | 0.14 |
ENSMUST00000047721.8
|
Rrp9
|
RRP9, small subunit (SSU) processome component, homolog (yeast) |
| chr1_-_171607321 | 0.14 |
ENSMUST00000111277.1
ENSMUST00000004827.7 |
Ly9
|
lymphocyte antigen 9 |
| chr1_+_92910805 | 0.13 |
ENSMUST00000179711.1
|
Rnpepl1
|
arginyl aminopeptidase (aminopeptidase B)-like 1 |
| chr15_-_79774383 | 0.13 |
ENSMUST00000069877.5
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr15_-_98004695 | 0.10 |
ENSMUST00000023123.8
|
Col2a1
|
collagen, type II, alpha 1 |
| chr11_-_70322520 | 0.10 |
ENSMUST00000019051.2
|
Alox12e
|
arachidonate lipoxygenase, epidermal |
| chr11_-_100441058 | 0.09 |
ENSMUST00000107399.2
ENSMUST00000092688.5 |
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr10_+_88201158 | 0.09 |
ENSMUST00000171151.2
|
Ccdc53
|
coiled-coil domain containing 53 |
| chr15_-_98004634 | 0.08 |
ENSMUST00000131560.1
ENSMUST00000088355.5 |
Col2a1
|
collagen, type II, alpha 1 |
| chr9_-_106476372 | 0.07 |
ENSMUST00000123555.1
ENSMUST00000125850.1 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr7_-_4532419 | 0.06 |
ENSMUST00000094897.4
|
Dnaaf3
|
dynein, axonemal assembly factor 3 |
| chr11_+_50131342 | 0.06 |
ENSMUST00000093138.6
ENSMUST00000101270.4 |
Tbc1d9b
|
TBC1 domain family, member 9B |
| chr11_+_66956620 | 0.05 |
ENSMUST00000150220.1
|
9130409J20Rik
|
RIKEN cDNA 9130409J20 gene |
| chr6_+_83078339 | 0.04 |
ENSMUST00000165164.2
ENSMUST00000092614.2 |
Pcgf1
|
polycomb group ring finger 1 |
| chr19_-_7241216 | 0.04 |
ENSMUST00000025675.9
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chrY_-_40391443 | 0.03 |
ENSMUST00000178865.1
|
Gm21801
|
predicted gene, 21801 |
| chrY_-_40457264 | 0.03 |
ENSMUST00000178452.1
|
Gm21914
|
predicted gene, 21914 |
| chr4_-_12087912 | 0.03 |
ENSMUST00000050686.3
|
Tmem67
|
transmembrane protein 67 |
| chr1_-_171607378 | 0.03 |
ENSMUST00000068878.7
|
Ly9
|
lymphocyte antigen 9 |
| chr10_+_79034723 | 0.02 |
ENSMUST00000082244.2
|
Olfr57
|
olfactory receptor 57 |
| chrY_+_32159845 | 0.01 |
ENSMUST00000178692.1
|
Gm21723
|
predicted gene, 21723 |
| chrY_+_32221125 | 0.01 |
ENSMUST00000179646.1
|
Gm21821
|
predicted gene, 21821 |
| chrY_+_32283545 | 0.01 |
ENSMUST00000177552.1
|
Gm21842
|
predicted gene, 21842 |
| chrY_-_30906508 | 0.01 |
ENSMUST00000177925.1
|
Gm21539
|
predicted gene, 21539 |
| chrY_-_31736484 | 0.01 |
ENSMUST00000177888.1
|
Gm21562
|
predicted gene, 21562 |
| chrY_-_31802731 | 0.01 |
ENSMUST00000180134.1
|
Gm21572
|
predicted gene, 21572 |
| chrY_-_32779761 | 0.01 |
ENSMUST00000178249.1
|
Gm21599
|
predicted gene, 21599 |
| chrY_-_33636191 | 0.01 |
ENSMUST00000177791.1
|
Gm21626
|
predicted gene, 21626 |
| chrY_-_33702857 | 0.01 |
ENSMUST00000178941.1
|
Gm21633
|
predicted gene, 21633 |
| chr11_+_70017085 | 0.01 |
ENSMUST00000108589.2
|
Dlg4
|
discs, large homolog 4 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ptf1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 1.0 | 3.0 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.7 | 109.6 | GO:0007586 | digestion(GO:0007586) |
| 0.6 | 1.9 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.5 | 1.4 | GO:2000412 | lymphocyte migration into lymphoid organs(GO:0097021) positive regulation of thymocyte migration(GO:2000412) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.5 | 1.4 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.4 | 0.8 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.4 | 1.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.3 | 1.0 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.3 | 1.2 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.3 | 0.5 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.2 | 0.7 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.2 | 0.9 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 1.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.5 | GO:0060857 | establishment of glial blood-brain barrier(GO:0060857) |
| 0.1 | 0.5 | GO:0060447 | peripheral nervous system axon regeneration(GO:0014012) bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.8 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 15.0 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 0.8 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.1 | 0.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 0.4 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.1 | 0.2 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.6 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.2 | GO:0046271 | coumarin metabolic process(GO:0009804) phenylpropanoid catabolic process(GO:0046271) |
| 0.1 | 0.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.4 | GO:1905049 | negative regulation of metallopeptidase activity(GO:1905049) |
| 0.1 | 1.4 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 1.0 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.1 | 0.2 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 1.0 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 18.2 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 3.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 6.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.2 | GO:0009814 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.4 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 1.0 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.3 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.3 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.2 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.8 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.2 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.8 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 1.2 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.6 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 15.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 18.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 16.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 101.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 2.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.5 | 112.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.5 | 1.5 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.3 | 2.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 1.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.3 | 1.4 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.2 | 5.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 0.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 0.8 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 1.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.3 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.7 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.2 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.1 | 0.4 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.4 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 0.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.2 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 3.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.5 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.8 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.6 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 2.9 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 1.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 23.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.7 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 26.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.6 | 7.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 3.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |