Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
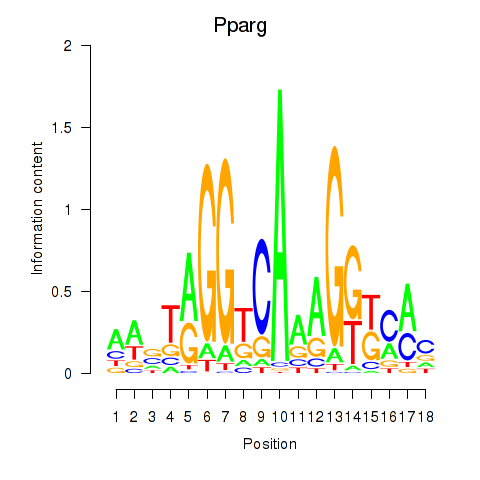
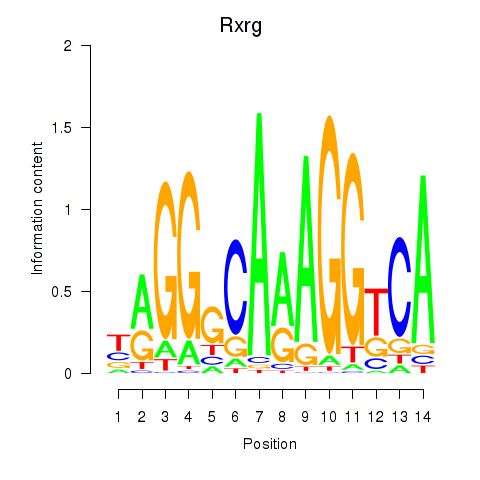
Results for Pparg_Rxrg
Z-value: 2.98
Transcription factors associated with Pparg_Rxrg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pparg
|
ENSMUSG00000000440.6 | peroxisome proliferator activated receptor gamma |
|
Rxrg
|
ENSMUSG00000015843.4 | retinoid X receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rxrg | mm10_v2_chr1_+_167598384_167598411 | 0.91 | 7.4e-15 | Click! |
| Pparg | mm10_v2_chr6_+_115422040_115422067 | 0.45 | 5.8e-03 | Click! |
Activity profile of Pparg_Rxrg motif
Sorted Z-values of Pparg_Rxrg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_46131888 | 67.12 |
ENSMUST00000043739.3
|
Elovl3
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
| chr17_-_46438471 | 59.99 |
ENSMUST00000087012.5
|
Slc22a7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chr19_+_39287074 | 52.47 |
ENSMUST00000003137.8
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr9_-_46235631 | 48.08 |
ENSMUST00000118649.1
|
Apoc3
|
apolipoprotein C-III |
| chr9_-_46235260 | 36.17 |
ENSMUST00000121916.1
ENSMUST00000034586.2 |
Apoc3
|
apolipoprotein C-III |
| chr15_-_82764176 | 34.84 |
ENSMUST00000055721.4
|
Cyp2d40
|
cytochrome P450, family 2, subfamily d, polypeptide 40 |
| chr3_+_138415484 | 25.60 |
ENSMUST00000161312.1
ENSMUST00000013458.8 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr3_+_138374121 | 21.87 |
ENSMUST00000171054.1
|
Adh6-ps1
|
alcohol dehydrogenase 6 (class V), pseudogene 1 |
| chr9_-_15301555 | 20.76 |
ENSMUST00000034414.8
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr15_+_82555087 | 20.00 |
ENSMUST00000068861.6
|
Cyp2d12
|
cytochrome P450, family 2, subfamily d, polypeptide 12 |
| chr2_+_173153048 | 19.35 |
ENSMUST00000029017.5
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr4_-_60501903 | 18.33 |
ENSMUST00000084548.4
ENSMUST00000103012.3 ENSMUST00000107499.3 |
Mup1
|
major urinary protein 1 |
| chr6_-_85820965 | 18.02 |
ENSMUST00000032074.3
|
Cml5
|
camello-like 5 |
| chr7_-_99695628 | 17.31 |
ENSMUST00000145381.1
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr4_-_62087261 | 17.28 |
ENSMUST00000107488.3
ENSMUST00000107472.1 ENSMUST00000084531.4 |
Mup3
|
major urinary protein 3 |
| chr4_-_62054112 | 17.15 |
ENSMUST00000074018.3
|
Mup20
|
major urinary protein 20 |
| chr4_-_60741275 | 17.04 |
ENSMUST00000117932.1
|
Mup12
|
major urinary protein 12 |
| chr7_-_99695572 | 16.07 |
ENSMUST00000137914.1
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr7_-_97417730 | 15.95 |
ENSMUST00000043077.7
|
Thrsp
|
thyroid hormone responsive |
| chr11_-_78422217 | 15.84 |
ENSMUST00000001122.5
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr19_+_36554661 | 15.56 |
ENSMUST00000169036.2
ENSMUST00000047247.5 |
Hectd2
|
HECT domain containing 2 |
| chr6_-_85869128 | 14.89 |
ENSMUST00000045008.7
|
Cml2
|
camello-like 2 |
| chr15_-_82620907 | 14.70 |
ENSMUST00000109515.1
|
Cyp2d34
|
cytochrome P450, family 2, subfamily d, polypeptide 34 |
| chr3_+_94693556 | 14.46 |
ENSMUST00000090848.3
ENSMUST00000173981.1 ENSMUST00000173849.1 ENSMUST00000174223.1 |
Selenbp2
|
selenium binding protein 2 |
| chr1_-_180195981 | 13.99 |
ENSMUST00000027766.6
ENSMUST00000161814.1 |
Adck3
|
aarF domain containing kinase 3 |
| chr5_+_114146525 | 13.97 |
ENSMUST00000102582.1
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr10_-_128960965 | 13.68 |
ENSMUST00000026398.3
|
Mettl7b
|
methyltransferase like 7B |
| chr19_-_40187277 | 13.44 |
ENSMUST00000051846.6
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr15_+_82452372 | 13.13 |
ENSMUST00000089129.5
|
Cyp2d9
|
cytochrome P450, family 2, subfamily d, polypeptide 9 |
| chr9_-_86695897 | 13.09 |
ENSMUST00000034989.8
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr17_-_32917048 | 12.60 |
ENSMUST00000054174.7
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr5_-_110286159 | 12.45 |
ENSMUST00000031472.5
|
Pxmp2
|
peroxisomal membrane protein 2 |
| chr12_-_30373358 | 12.44 |
ENSMUST00000021004.7
|
Sntg2
|
syntrophin, gamma 2 |
| chr6_+_72636244 | 12.23 |
ENSMUST00000101278.2
|
Gm15401
|
predicted gene 15401 |
| chr7_-_99695809 | 12.05 |
ENSMUST00000107086.2
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr19_-_8405060 | 11.92 |
ENSMUST00000064507.5
ENSMUST00000120540.1 ENSMUST00000096269.4 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr17_-_34882004 | 11.71 |
ENSMUST00000152417.1
ENSMUST00000146299.1 |
C2
Gm20547
|
complement component 2 (within H-2S) predicted gene 20547 |
| chr9_+_46240696 | 11.43 |
ENSMUST00000034585.6
|
Apoa4
|
apolipoprotein A-IV |
| chr10_+_127801145 | 11.34 |
ENSMUST00000071646.1
|
Rdh16
|
retinol dehydrogenase 16 |
| chr17_-_32917320 | 11.34 |
ENSMUST00000179434.1
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr2_-_32704123 | 11.33 |
ENSMUST00000127812.1
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr13_-_41847626 | 11.33 |
ENSMUST00000121404.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr13_-_93637961 | 10.99 |
ENSMUST00000099309.4
|
Bhmt
|
betaine-homocysteine methyltransferase |
| chr11_+_75468040 | 10.97 |
ENSMUST00000043598.7
ENSMUST00000108435.1 |
Tlcd2
|
TLC domain containing 2 |
| chr9_-_22002599 | 10.92 |
ENSMUST00000115336.2
ENSMUST00000044926.5 |
Ccdc151
|
coiled-coil domain containing 151 |
| chr18_-_61911783 | 10.90 |
ENSMUST00000049378.8
ENSMUST00000166783.1 |
Ablim3
|
actin binding LIM protein family, member 3 |
| chr7_+_140763739 | 10.87 |
ENSMUST00000026552.7
|
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr7_+_140835018 | 10.69 |
ENSMUST00000106050.1
ENSMUST00000026554.4 |
Urah
|
urate (5-hydroxyiso-) hydrolase |
| chr3_+_130617448 | 10.66 |
ENSMUST00000166187.1
ENSMUST00000072271.6 |
Etnppl
|
ethanolamine phosphate phospholyase |
| chr10_+_87859062 | 10.62 |
ENSMUST00000095360.4
|
Igf1
|
insulin-like growth factor 1 |
| chr18_-_38866702 | 10.57 |
ENSMUST00000115582.1
|
Fgf1
|
fibroblast growth factor 1 |
| chr11_+_70054334 | 10.48 |
ENSMUST00000018699.6
ENSMUST00000108585.2 |
Asgr1
|
asialoglycoprotein receptor 1 |
| chr2_-_25500613 | 10.41 |
ENSMUST00000040042.4
|
C8g
|
complement component 8, gamma polypeptide |
| chr19_+_39007019 | 10.33 |
ENSMUST00000025966.4
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr11_-_77894096 | 10.33 |
ENSMUST00000017597.4
|
Pipox
|
pipecolic acid oxidase |
| chr6_+_72598475 | 10.06 |
ENSMUST00000070597.6
ENSMUST00000176364.1 ENSMUST00000176168.1 |
Retsat
|
retinol saturase (all trans retinol 13,14 reductase) |
| chr19_+_12633303 | 10.05 |
ENSMUST00000044976.5
|
Glyat
|
glycine-N-acyltransferase |
| chr4_-_62150810 | 9.97 |
ENSMUST00000077719.3
|
Mup21
|
major urinary protein 21 |
| chr19_+_39992424 | 9.91 |
ENSMUST00000049178.2
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr17_-_73950172 | 9.85 |
ENSMUST00000024866.4
|
Xdh
|
xanthine dehydrogenase |
| chr9_+_46269069 | 9.62 |
ENSMUST00000034584.3
|
Apoa5
|
apolipoprotein A-V |
| chr10_+_127849917 | 9.58 |
ENSMUST00000077530.2
|
Rdh19
|
retinol dehydrogenase 19 |
| chr1_-_132139666 | 9.54 |
ENSMUST00000027697.5
|
Cdk18
|
cyclin-dependent kinase 18 |
| chr1_-_132139605 | 9.51 |
ENSMUST00000112362.2
|
Cdk18
|
cyclin-dependent kinase 18 |
| chr13_-_41847599 | 9.48 |
ENSMUST00000179758.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr8_+_104847061 | 9.38 |
ENSMUST00000055052.5
|
Ces2c
|
carboxylesterase 2C |
| chr19_+_39510844 | 9.31 |
ENSMUST00000025968.4
|
Cyp2c39
|
cytochrome P450, family 2, subfamily c, polypeptide 39 |
| chr12_-_104044431 | 9.23 |
ENSMUST00000043915.3
|
Serpina12
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| chr6_+_125320633 | 8.97 |
ENSMUST00000176655.1
ENSMUST00000176110.1 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr15_+_9335550 | 8.97 |
ENSMUST00000072403.6
|
Ugt3a2
|
UDP glycosyltransferases 3 family, polypeptide A2 |
| chr3_+_130617645 | 8.95 |
ENSMUST00000163620.1
|
Etnppl
|
ethanolamine phosphate phospholyase |
| chr15_+_9279829 | 8.90 |
ENSMUST00000022861.8
|
Ugt3a1
|
UDP glycosyltransferases 3 family, polypeptide A1 |
| chr2_+_160888101 | 8.84 |
ENSMUST00000109455.2
ENSMUST00000040872.6 |
Lpin3
|
lipin 3 |
| chr5_-_130024280 | 8.81 |
ENSMUST00000161640.1
ENSMUST00000161884.1 ENSMUST00000161094.1 |
Asl
|
argininosuccinate lyase |
| chr2_+_160888156 | 8.81 |
ENSMUST00000109457.2
|
Lpin3
|
lipin 3 |
| chr9_-_103228420 | 8.62 |
ENSMUST00000126359.1
|
Trf
|
transferrin |
| chr6_-_85933379 | 8.61 |
ENSMUST00000162660.1
|
Nat8b
|
N-acetyltransferase 8B |
| chr14_-_79662148 | 8.53 |
ENSMUST00000022603.7
|
Lect1
|
leukocyte cell derived chemotaxin 1 |
| chr2_+_126556128 | 8.50 |
ENSMUST00000141482.2
|
Slc27a2
|
solute carrier family 27 (fatty acid transporter), member 2 |
| chr4_-_134372529 | 8.47 |
ENSMUST00000030643.2
|
Extl1
|
exostoses (multiple)-like 1 |
| chr14_-_79662084 | 8.47 |
ENSMUST00000165835.1
|
Lect1
|
leukocyte cell derived chemotaxin 1 |
| chr10_+_127759780 | 8.45 |
ENSMUST00000128247.1
|
RP23-386P10.11
|
Protein Rdh9 |
| chr19_-_39649046 | 8.43 |
ENSMUST00000067328.6
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr2_+_25080316 | 8.41 |
ENSMUST00000044078.3
ENSMUST00000114380.2 |
Entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr1_+_72824482 | 8.41 |
ENSMUST00000047328.4
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr17_-_34882094 | 8.28 |
ENSMUST00000025230.8
|
C2
|
complement component 2 (within H-2S) |
| chr8_+_13026024 | 8.19 |
ENSMUST00000033820.3
|
F7
|
coagulation factor VII |
| chr7_-_25477607 | 8.16 |
ENSMUST00000098669.1
ENSMUST00000098668.1 ENSMUST00000098666.2 |
Ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr6_+_83156401 | 8.13 |
ENSMUST00000032106.4
|
1700003E16Rik
|
RIKEN cDNA 1700003E16 gene |
| chr7_+_44207307 | 7.89 |
ENSMUST00000077354.4
|
Klk1b4
|
kallikrein 1-related pepidase b4 |
| chrX_+_150594420 | 7.89 |
ENSMUST00000112713.2
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr9_-_106476104 | 7.89 |
ENSMUST00000156426.1
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr16_-_19200350 | 7.88 |
ENSMUST00000103749.2
|
Iglc2
|
immunoglobulin lambda constant 2 |
| chr7_-_105600103 | 7.87 |
ENSMUST00000033185.8
|
Hpx
|
hemopexin |
| chr7_-_14438538 | 7.69 |
ENSMUST00000168252.2
|
2810007J24Rik
|
RIKEN cDNA 2810007J24 gene |
| chr11_-_70015346 | 7.63 |
ENSMUST00000018718.7
ENSMUST00000102574.3 |
Acadvl
|
acyl-Coenzyme A dehydrogenase, very long chain |
| chr4_-_103215147 | 7.61 |
ENSMUST00000150285.1
|
Slc35d1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
| chr15_-_82394022 | 7.60 |
ENSMUST00000170255.1
|
Cyp2d11
|
cytochrome P450, family 2, subfamily d, polypeptide 11 |
| chr19_-_44029201 | 7.51 |
ENSMUST00000026211.8
|
Cyp2c44
|
cytochrome P450, family 2, subfamily c, polypeptide 44 |
| chr17_-_34028044 | 7.43 |
ENSMUST00000045467.7
ENSMUST00000114303.3 |
H2-Ke6
|
H2-K region expressed gene 6 |
| chr6_+_129533183 | 7.30 |
ENSMUST00000032264.6
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr11_-_113710017 | 7.22 |
ENSMUST00000018871.1
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr10_+_128254131 | 7.21 |
ENSMUST00000060782.3
|
Apon
|
apolipoprotein N |
| chr10_+_128790903 | 7.21 |
ENSMUST00000026411.6
|
Mmp19
|
matrix metallopeptidase 19 |
| chr13_+_3634032 | 7.12 |
ENSMUST00000042288.6
|
Asb13
|
ankyrin repeat and SOCS box-containing 13 |
| chr8_-_110039330 | 7.07 |
ENSMUST00000109222.2
|
Chst4
|
carbohydrate (chondroitin 6/keratan) sulfotransferase 4 |
| chr2_+_162987502 | 6.99 |
ENSMUST00000117123.1
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr5_+_120476522 | 6.93 |
ENSMUST00000066540.7
|
Sds
|
serine dehydratase |
| chr7_-_68275098 | 6.93 |
ENSMUST00000135564.1
|
Gm16157
|
predicted gene 16157 |
| chr2_+_162987330 | 6.88 |
ENSMUST00000018012.7
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr4_+_148602527 | 6.88 |
ENSMUST00000105701.2
ENSMUST00000052060.6 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr2_+_102706356 | 6.87 |
ENSMUST00000123759.1
ENSMUST00000111212.1 ENSMUST00000005220.4 |
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr15_-_82690499 | 6.86 |
ENSMUST00000100380.3
|
Cyp2d37-ps
|
cytochrome P450, family 2, subfamily d, polypeptide 37, pseudogene |
| chr5_+_130448801 | 6.86 |
ENSMUST00000111288.2
|
Caln1
|
calneuron 1 |
| chr13_+_91741507 | 6.73 |
ENSMUST00000022120.4
|
Acot12
|
acyl-CoA thioesterase 12 |
| chr4_-_59960659 | 6.70 |
ENSMUST00000075973.2
|
Mup4
|
major urinary protein 4 |
| chr19_+_12633507 | 6.69 |
ENSMUST00000119960.1
|
Glyat
|
glycine-N-acyltransferase |
| chr5_+_92571544 | 6.62 |
ENSMUST00000082382.7
|
Fam47e
|
family with sequence similarity 47, member E |
| chr6_-_85820936 | 6.59 |
ENSMUST00000174143.1
|
Gm11128
|
predicted gene 11128 |
| chr15_-_76090013 | 6.59 |
ENSMUST00000019516.4
|
Nrbp2
|
nuclear receptor binding protein 2 |
| chr4_-_42756543 | 6.58 |
ENSMUST00000102957.3
|
Ccl19
|
chemokine (C-C motif) ligand 19 |
| chr10_-_127370535 | 6.53 |
ENSMUST00000026472.8
|
Inhbc
|
inhibin beta-C |
| chr4_-_141623799 | 6.52 |
ENSMUST00000038661.7
|
Slc25a34
|
solute carrier family 25, member 34 |
| chr16_+_5007283 | 6.50 |
ENSMUST00000184439.1
|
Smim22
|
small integral membrane protein 22 |
| chr10_+_128267997 | 6.49 |
ENSMUST00000050901.2
|
Apof
|
apolipoprotein F |
| chr10_+_3540240 | 6.49 |
ENSMUST00000019896.4
|
Iyd
|
iodotyrosine deiodinase |
| chr15_-_77533312 | 6.44 |
ENSMUST00000062562.5
|
Apol7c
|
apolipoprotein L 7c |
| chr1_-_180193475 | 6.43 |
ENSMUST00000160482.1
ENSMUST00000170472.1 |
Adck3
|
aarF domain containing kinase 3 |
| chr7_+_63916857 | 6.38 |
ENSMUST00000177638.1
|
E030018B13Rik
|
RIKEN cDNA E030018B13 gene |
| chr9_-_106476590 | 6.33 |
ENSMUST00000112479.2
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr5_+_92571477 | 6.31 |
ENSMUST00000176621.1
ENSMUST00000175974.1 ENSMUST00000131166.2 ENSMUST00000176448.1 |
Fam47e
|
family with sequence similarity 47, member E |
| chr9_+_46012810 | 6.24 |
ENSMUST00000126865.1
|
Sik3
|
SIK family kinase 3 |
| chr1_-_180195902 | 6.19 |
ENSMUST00000161746.1
|
Adck3
|
aarF domain containing kinase 3 |
| chr15_+_3270767 | 6.19 |
ENSMUST00000082424.4
ENSMUST00000159158.1 ENSMUST00000159216.1 ENSMUST00000160311.1 |
Sepp1
|
selenoprotein P, plasma, 1 |
| chr18_+_74779190 | 6.16 |
ENSMUST00000041053.9
|
Acaa2
|
acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) |
| chr6_+_121343052 | 6.09 |
ENSMUST00000166457.1
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr11_+_102761402 | 6.07 |
ENSMUST00000103081.4
ENSMUST00000068150.5 |
Adam11
|
a disintegrin and metallopeptidase domain 11 |
| chr19_+_11536769 | 6.03 |
ENSMUST00000025581.6
|
Ms4a4d
|
membrane-spanning 4-domains, subfamily A, member 4D |
| chr7_+_43444104 | 6.03 |
ENSMUST00000004729.3
|
Etfb
|
electron transferring flavoprotein, beta polypeptide |
| chr13_-_41828418 | 6.00 |
ENSMUST00000137905.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr1_+_181051232 | 5.98 |
ENSMUST00000036819.6
|
9130409I23Rik
|
RIKEN cDNA 9130409I23 gene |
| chr11_-_95041335 | 5.95 |
ENSMUST00000038431.7
|
Pdk2
|
pyruvate dehydrogenase kinase, isoenzyme 2 |
| chr9_-_103288224 | 5.94 |
ENSMUST00000123530.1
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr5_-_45450143 | 5.94 |
ENSMUST00000154962.1
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr4_+_141242850 | 5.93 |
ENSMUST00000138096.1
ENSMUST00000006618.2 ENSMUST00000125392.1 |
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr9_+_108662098 | 5.92 |
ENSMUST00000035222.5
|
Slc25a20
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 |
| chr2_+_30266721 | 5.91 |
ENSMUST00000113645.1
ENSMUST00000133877.1 ENSMUST00000139719.1 ENSMUST00000113643.1 ENSMUST00000150695.1 |
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr7_-_12998172 | 5.90 |
ENSMUST00000120903.1
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr9_-_106476372 | 5.85 |
ENSMUST00000123555.1
ENSMUST00000125850.1 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr9_-_103288290 | 5.84 |
ENSMUST00000035163.3
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr16_-_93929512 | 5.82 |
ENSMUST00000177648.1
|
Cldn14
|
claudin 14 |
| chr9_-_55512156 | 5.81 |
ENSMUST00000034866.8
|
Etfa
|
electron transferring flavoprotein, alpha polypeptide |
| chr15_-_82407187 | 5.79 |
ENSMUST00000072776.3
|
Cyp2d10
|
cytochrome P450, family 2, subfamily d, polypeptide 10 |
| chr4_+_148130883 | 5.74 |
ENSMUST00000084129.2
|
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr11_+_115462464 | 5.66 |
ENSMUST00000106532.3
ENSMUST00000092445.5 ENSMUST00000153466.1 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr7_-_12998140 | 5.64 |
ENSMUST00000032539.7
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr6_-_72235559 | 5.64 |
ENSMUST00000042646.7
|
Atoh8
|
atonal homolog 8 (Drosophila) |
| chr12_+_108851122 | 5.63 |
ENSMUST00000057026.8
|
Slc25a47
|
solute carrier family 25, member 47 |
| chr7_-_30944017 | 5.62 |
ENSMUST00000062620.7
|
Hamp
|
hepcidin antimicrobial peptide |
| chr11_+_101367542 | 5.62 |
ENSMUST00000019469.2
|
G6pc
|
glucose-6-phosphatase, catalytic |
| chr8_-_25038875 | 5.57 |
ENSMUST00000084031.4
|
Htra4
|
HtrA serine peptidase 4 |
| chr5_-_38480131 | 5.54 |
ENSMUST00000143758.1
ENSMUST00000067886.5 |
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr15_-_83170168 | 5.52 |
ENSMUST00000162834.1
|
Cyb5r3
|
cytochrome b5 reductase 3 |
| chr19_+_4711153 | 5.51 |
ENSMUST00000008991.6
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr2_-_103485068 | 5.47 |
ENSMUST00000111168.3
|
Cat
|
catalase |
| chr1_+_139501692 | 5.47 |
ENSMUST00000027615.5
|
F13b
|
coagulation factor XIII, beta subunit |
| chr2_-_160872829 | 5.44 |
ENSMUST00000176141.1
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr17_-_34804546 | 5.40 |
ENSMUST00000025223.8
|
Cyp21a1
|
cytochrome P450, family 21, subfamily a, polypeptide 1 |
| chr8_+_104733997 | 5.40 |
ENSMUST00000034346.8
ENSMUST00000164182.2 |
Ces2a
|
carboxylesterase 2A |
| chr15_+_7129557 | 5.39 |
ENSMUST00000067190.5
ENSMUST00000164529.1 |
Lifr
|
leukemia inhibitory factor receptor |
| chr4_-_155398059 | 5.34 |
ENSMUST00000030925.2
|
Gabrd
|
gamma-aminobutyric acid (GABA) A receptor, subunit delta |
| chr14_+_66140919 | 5.31 |
ENSMUST00000022620.9
|
Chrna2
|
cholinergic receptor, nicotinic, alpha polypeptide 2 (neuronal) |
| chr10_+_79890853 | 5.30 |
ENSMUST00000061653.7
|
Cfd
|
complement factor D (adipsin) |
| chr18_+_20944607 | 5.30 |
ENSMUST00000050004.1
|
Rnf125
|
ring finger protein 125 |
| chr5_-_45450121 | 5.29 |
ENSMUST00000127562.1
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr1_-_180245927 | 5.28 |
ENSMUST00000010753.7
|
Psen2
|
presenilin 2 |
| chr7_-_90129339 | 5.25 |
ENSMUST00000181189.1
|
2310010J17Rik
|
RIKEN cDNA 2310010J17 gene |
| chr9_+_46012822 | 5.25 |
ENSMUST00000120463.2
ENSMUST00000120247.1 |
Sik3
|
SIK family kinase 3 |
| chr6_+_82052307 | 5.23 |
ENSMUST00000149023.1
|
Eva1a
|
eva-1 homolog A (C. elegans) |
| chr19_-_6921804 | 5.22 |
ENSMUST00000025906.4
|
Esrra
|
estrogen related receptor, alpha |
| chr16_+_5007306 | 5.21 |
ENSMUST00000178155.2
ENSMUST00000184256.1 ENSMUST00000185147.1 |
Smim22
|
small integral membrane protein 22 |
| chr14_+_27622433 | 5.19 |
ENSMUST00000090302.5
|
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr2_-_91636394 | 5.15 |
ENSMUST00000111335.1
ENSMUST00000028681.8 |
F2
|
coagulation factor II |
| chr13_-_41847482 | 5.05 |
ENSMUST00000072012.3
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr3_+_118562129 | 5.05 |
ENSMUST00000039177.7
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr2_+_122765377 | 5.04 |
ENSMUST00000124460.1
ENSMUST00000147475.1 |
Sqrdl
|
sulfide quinone reductase-like (yeast) |
| chr1_-_162898665 | 5.04 |
ENSMUST00000111510.1
ENSMUST00000045902.6 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr2_-_91195097 | 5.04 |
ENSMUST00000002177.2
ENSMUST00000111354.1 |
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr2_-_91195035 | 5.03 |
ENSMUST00000111356.1
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr18_-_38929148 | 5.01 |
ENSMUST00000134864.1
|
Fgf1
|
fibroblast growth factor 1 |
| chr11_+_120530688 | 5.01 |
ENSMUST00000026119.7
|
Gcgr
|
glucagon receptor |
| chr1_+_78511865 | 4.96 |
ENSMUST00000012331.6
|
Mogat1
|
monoacylglycerol O-acyltransferase 1 |
| chr17_-_45686120 | 4.94 |
ENSMUST00000143907.1
ENSMUST00000127065.1 |
Tmem63b
|
transmembrane protein 63b |
| chr15_-_82380179 | 4.94 |
ENSMUST00000023083.7
|
Cyp2d22
|
cytochrome P450, family 2, subfamily d, polypeptide 22 |
| chr1_-_180245757 | 4.92 |
ENSMUST00000111104.1
|
Psen2
|
presenilin 2 |
| chr15_-_77447444 | 4.91 |
ENSMUST00000089469.5
|
Apol7b
|
apolipoprotein L 7b |
| chr1_+_87574016 | 4.89 |
ENSMUST00000166259.1
ENSMUST00000172222.1 ENSMUST00000163606.1 |
Neu2
|
neuraminidase 2 |
| chr6_-_48445678 | 4.89 |
ENSMUST00000114556.1
|
Zfp467
|
zinc finger protein 467 |
| chr10_-_89506631 | 4.87 |
ENSMUST00000058126.8
ENSMUST00000105296.2 |
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr4_-_130279205 | 4.86 |
ENSMUST00000120126.2
|
Serinc2
|
serine incorporator 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pparg_Rxrg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 21.1 | 84.3 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 11.4 | 45.4 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 7.7 | 23.1 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 6.7 | 20.1 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 6.4 | 25.6 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 6.1 | 18.3 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 5.9 | 71.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 4.8 | 14.4 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 4.7 | 14.0 | GO:2001293 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 3.8 | 11.4 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) multicellular organism lipid catabolic process(GO:0044240) |
| 3.8 | 11.3 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 3.7 | 111.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 3.6 | 18.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 3.5 | 104.1 | GO:0080184 | response to phenylpropanoid(GO:0080184) |
| 3.4 | 10.3 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 3.4 | 126.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 3.1 | 12.5 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 2.9 | 11.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 2.9 | 17.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 2.8 | 8.5 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 2.7 | 8.2 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 2.5 | 10.1 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 2.4 | 17.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 2.4 | 7.2 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 2.4 | 7.2 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 2.4 | 7.1 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 2.2 | 8.7 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) fatty-acyl-CoA catabolic process(GO:0036115) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 2.1 | 18.7 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 2.0 | 8.2 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 2.0 | 4.1 | GO:0035963 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 2.0 | 6.0 | GO:0042335 | cuticle development(GO:0042335) |
| 2.0 | 14.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 1.9 | 7.8 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.9 | 13.1 | GO:0006108 | malate metabolic process(GO:0006108) regulation of NADP metabolic process(GO:1902031) |
| 1.8 | 7.3 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 1.8 | 10.7 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 1.8 | 10.6 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 1.7 | 9.9 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 1.6 | 4.9 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 1.6 | 11.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.6 | 7.9 | GO:0015886 | heme transport(GO:0015886) |
| 1.6 | 31.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 1.6 | 4.7 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 1.5 | 7.7 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 1.5 | 6.2 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 1.5 | 5.9 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 1.5 | 4.4 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 1.4 | 5.7 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 1.4 | 20.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.4 | 7.1 | GO:0034627 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 1.4 | 5.6 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 1.4 | 4.2 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.4 | 2.7 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 1.4 | 4.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 1.3 | 4.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 1.3 | 10.4 | GO:0098707 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 1.3 | 5.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 1.2 | 11.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 1.2 | 14.9 | GO:0046415 | urate metabolic process(GO:0046415) |
| 1.2 | 6.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 1.2 | 4.6 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 1.1 | 15.8 | GO:0015747 | urate transport(GO:0015747) |
| 1.1 | 6.7 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 1.1 | 3.3 | GO:2000584 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 1.1 | 4.3 | GO:1902728 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 1.1 | 5.4 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 1.1 | 10.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.1 | 3.2 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 1.1 | 3.2 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 1.0 | 5.2 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 1.0 | 7.3 | GO:0030242 | pexophagy(GO:0030242) |
| 1.0 | 3.1 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 1.0 | 8.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 1.0 | 5.1 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 1.0 | 4.0 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 1.0 | 2.9 | GO:0031038 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 1.0 | 6.9 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.0 | 8.8 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 1.0 | 1.9 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 1.0 | 7.6 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.9 | 2.8 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.9 | 2.8 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.9 | 2.8 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.9 | 3.7 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.9 | 2.8 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.9 | 35.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.9 | 5.4 | GO:1903275 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.9 | 10.7 | GO:0009133 | nucleoside diphosphate biosynthetic process(GO:0009133) |
| 0.9 | 7.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.9 | 7.0 | GO:0035437 | maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.9 | 3.5 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.9 | 3.5 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.8 | 6.7 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.8 | 2.5 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.8 | 5.0 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.8 | 1.7 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.8 | 7.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.8 | 4.8 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.8 | 11.8 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.8 | 2.3 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.8 | 3.1 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.8 | 4.5 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.8 | 5.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.7 | 2.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.7 | 8.5 | GO:0009650 | UV protection(GO:0009650) |
| 0.7 | 4.9 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.7 | 2.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.7 | 0.7 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.7 | 2.1 | GO:1903048 | regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.7 | 2.7 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.7 | 4.7 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.6 | 8.4 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.6 | 3.8 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.6 | 1.9 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.6 | 2.5 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.6 | 7.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.6 | 1.8 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.6 | 7.1 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.6 | 4.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.6 | 10.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.6 | 4.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.6 | 2.3 | GO:0046618 | drug export(GO:0046618) |
| 0.6 | 5.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.6 | 1.2 | GO:0090244 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.6 | 5.7 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.6 | 2.8 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.6 | 2.3 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.6 | 3.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.6 | 9.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.5 | 3.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.5 | 2.7 | GO:1903679 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.5 | 2.7 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.5 | 2.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.5 | 2.7 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.5 | 1.6 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.5 | 3.6 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.5 | 1.5 | GO:0001193 | maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.5 | 6.5 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.5 | 1.5 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.5 | 5.4 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.5 | 1.5 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.5 | 1.5 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.5 | 2.9 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.5 | 4.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.5 | 3.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.5 | 0.9 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.5 | 3.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.5 | 5.5 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.4 | 2.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.4 | 1.8 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.4 | 0.9 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.4 | 1.3 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.4 | 2.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.4 | 1.7 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.4 | 2.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.4 | 0.8 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.4 | 2.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.4 | 5.4 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.4 | 1.2 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.4 | 4.1 | GO:0060283 | receptor guanylyl cyclase signaling pathway(GO:0007168) negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.4 | 2.8 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.4 | 2.4 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.4 | 2.0 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.4 | 1.6 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.4 | 1.2 | GO:1902605 | heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.4 | 2.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.4 | 1.2 | GO:0061026 | septum secundum development(GO:0003285) embryonic heart tube anterior/posterior pattern specification(GO:0035054) cardiac muscle tissue regeneration(GO:0061026) negative regulation of connective tissue replacement(GO:1905204) |
| 0.4 | 8.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.4 | 3.8 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.4 | 8.7 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.4 | 2.6 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.4 | 1.9 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.4 | 1.8 | GO:2000157 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.4 | 2.9 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.4 | 1.8 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.4 | 4.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 1.0 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.3 | 1.4 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.3 | 4.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.3 | 3.7 | GO:0006983 | ER overload response(GO:0006983) |
| 0.3 | 3.0 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.3 | 1.3 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.3 | 7.6 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.3 | 5.2 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.3 | 1.6 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.3 | 6.4 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.3 | 2.2 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.3 | 3.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.3 | 0.9 | GO:0003360 | brainstem development(GO:0003360) |
| 0.3 | 2.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.3 | 1.9 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.3 | 1.2 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.3 | 1.5 | GO:0097527 | microglial cell activation involved in immune response(GO:0002282) positive regulation of interferon-alpha biosynthetic process(GO:0045356) necroptotic signaling pathway(GO:0097527) |
| 0.3 | 1.2 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.3 | 1.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.3 | 1.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.3 | 1.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.3 | 2.7 | GO:0072641 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.3 | 11.5 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.3 | 0.9 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.3 | 1.8 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.3 | 1.2 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.3 | 4.0 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.3 | 7.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.3 | 1.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.3 | 1.1 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.3 | 0.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.3 | 2.7 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.3 | 0.8 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.3 | 2.1 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.3 | 1.3 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.3 | 3.9 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) |
| 0.3 | 1.0 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.3 | 1.8 | GO:0015684 | ferrous iron transport(GO:0015684) |
| 0.3 | 4.0 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.3 | 3.3 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.3 | 1.3 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.2 | 0.7 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.2 | 0.7 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.2 | 1.2 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.2 | 0.7 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.2 | 1.0 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.2 | 1.0 | GO:1990144 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) intrinsic apoptotic signaling pathway in response to hypoxia(GO:1990144) |
| 0.2 | 0.7 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.2 | 3.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 2.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.2 | 1.2 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.2 | 5.0 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.2 | 3.8 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.2 | 0.7 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.2 | 0.9 | GO:0015800 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) acidic amino acid transport(GO:0015800) |
| 0.2 | 1.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 0.9 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) myotome development(GO:0061055) |
| 0.2 | 5.4 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.2 | 2.7 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 10.0 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.2 | 0.9 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 | 2.4 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.2 | 7.7 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.2 | 0.8 | GO:0070162 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 1.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 0.6 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.2 | 1.2 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.2 | 1.8 | GO:0008228 | opsonization(GO:0008228) |
| 0.2 | 6.6 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.2 | 0.6 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.2 | 0.4 | GO:0071865 | regulation of apoptotic process in bone marrow(GO:0071865) negative regulation of apoptotic process in bone marrow(GO:0071866) |
| 0.2 | 0.8 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.2 | 1.0 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.2 | 1.3 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.2 | 1.5 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.2 | 1.9 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.2 | 4.8 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.2 | 2.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.8 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.2 | 0.7 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.2 | 0.9 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.2 | 1.5 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 1.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 | 0.9 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.2 | 1.0 | GO:0071838 | cell proliferation in bone marrow(GO:0071838) |
| 0.2 | 0.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 0.5 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.2 | 0.2 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.2 | 2.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.2 | 0.7 | GO:0061386 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 1.2 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.2 | 1.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.2 | 0.7 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 2.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 0.5 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.2 | 0.5 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 1.0 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.2 | 0.6 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.2 | 2.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 | 2.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.2 | 1.6 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.2 | 0.2 | GO:0003134 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.2 | 2.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 1.4 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.2 | 1.3 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 0.2 | 0.8 | GO:0060744 | positive regulation of exit from mitosis(GO:0031536) thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.2 | 0.6 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.2 | 0.8 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 2.8 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.2 | 0.9 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 0.9 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.2 | 2.6 | GO:0014823 | response to activity(GO:0014823) |
| 0.2 | 1.7 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.2 | 1.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.2 | 0.9 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.2 | 0.9 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 0.6 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.2 | 0.6 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.2 | 3.6 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 9.3 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 3.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 6.2 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 0.7 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 0.3 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.8 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 1.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 1.0 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.4 | GO:1900114 | spermatid nucleus elongation(GO:0007290) positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.1 | 5.7 | GO:0006641 | triglyceride metabolic process(GO:0006641) |
| 0.1 | 1.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.4 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.1 | 4.5 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 0.9 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 2.2 | GO:0071380 | cellular response to prostaglandin stimulus(GO:0071379) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 3.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 1.1 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 1.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 1.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 2.2 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 2.6 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.1 | 0.6 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 2.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 11.1 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 8.1 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 0.4 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 1.9 | GO:0070861 | regulation of protein exit from endoplasmic reticulum(GO:0070861) |
| 0.1 | 3.0 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 0.4 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.9 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 1.0 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.1 | 1.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 4.7 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.7 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.1 | 0.7 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 0.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 3.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 0.4 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.1 | 0.3 | GO:0072720 | response to sorbitol(GO:0072708) response to dithiothreitol(GO:0072720) |
| 0.1 | 0.8 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.1 | 0.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 1.0 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.1 | 2.3 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.1 | 0.3 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.1 | 0.6 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 0.3 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.7 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.7 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 1.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.3 | GO:2000864 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 0.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.0 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 1.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 4.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 3.4 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.2 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.1 | 3.7 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.5 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.9 | GO:1905214 | regulation of mRNA binding(GO:1902415) positive regulation of mRNA binding(GO:1902416) regulation of RNA binding(GO:1905214) positive regulation of RNA binding(GO:1905216) |
| 0.1 | 5.4 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 1.5 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 2.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.8 | GO:0097576 | autophagosome maturation(GO:0097352) vacuole fusion(GO:0097576) |
| 0.1 | 0.7 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 3.0 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 0.7 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 1.9 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 3.3 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 0.8 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) cristae formation(GO:0042407) |
| 0.1 | 5.2 | GO:0007632 | visual behavior(GO:0007632) |
| 0.1 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.2 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.1 | 1.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.2 | GO:0042414 | epinephrine metabolic process(GO:0042414) response to norepinephrine(GO:0071873) |
| 0.1 | 0.5 | GO:0060460 | left lung development(GO:0060459) left lung morphogenesis(GO:0060460) |
| 0.1 | 0.5 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 0.5 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 0.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) peptidyl-histidine modification(GO:0018202) |
| 0.1 | 1.3 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 2.9 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.5 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.2 | GO:1902915 | progesterone receptor signaling pathway(GO:0050847) negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 1.0 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.2 | GO:0072733 | regulation of retinal cell programmed cell death(GO:0046668) response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 1.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 2.7 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 0.3 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 | 0.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.2 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 5.1 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.1 | 2.6 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.1 | 1.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 0.8 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 2.3 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.1 | 0.5 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.2 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 2.9 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 0.7 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 5.8 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 0.4 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 5.4 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 2.0 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 1.2 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 2.1 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 0.2 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.1 | 9.4 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.1 | 0.7 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 1.0 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 1.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.2 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 4.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0000436 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) carbon catabolite activation of transcription(GO:0045991) positive regulation of transcription by glucose(GO:0046016) |
| 0.0 | 0.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.4 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.2 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.3 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.0 | 0.9 | GO:0051931 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.4 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 1.2 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 6.3 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.6 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.5 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.6 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.4 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.5 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.5 | GO:1901571 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 0.0 | 3.0 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.2 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.0 | 0.7 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.6 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.4 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.0 | GO:0050904 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) diapedesis(GO:0050904) |
| 0.0 | 1.8 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.6 | GO:0060487 | lung cell differentiation(GO:0060479) lung epithelial cell differentiation(GO:0060487) |
| 0.0 | 0.6 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 1.3 | GO:0050679 | positive regulation of epithelial cell proliferation(GO:0050679) |
| 0.0 | 0.3 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.3 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 0.2 | GO:0071554 | cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 0.5 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.4 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.0 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.5 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.3 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 83.8 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 4.3 | 17.1 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 2.6 | 7.9 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 2.2 | 8.9 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 2.0 | 5.9 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 1.8 | 20.2 | GO:0042627 | chylomicron(GO:0042627) |
| 1.2 | 10.6 | GO:0042567 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) insulin-like growth factor ternary complex(GO:0042567) |
| 1.1 | 10.9 | GO:0097433 | dense body(GO:0097433) |
| 1.0 | 5.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 1.0 | 31.0 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 1.0 | 3.0 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 1.0 | 6.8 | GO:0008091 | spectrin(GO:0008091) |
| 1.0 | 5.7 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.9 | 10.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.9 | 3.6 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.8 | 69.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.8 | 3.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.8 | 2.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.8 | 3.8 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.7 | 2.2 | GO:0044317 | rod spherule(GO:0044317) |
| 0.7 | 6.5 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.7 | 108.7 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.6 | 4.5 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.6 | 8.8 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.6 | 2.8 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.6 | 3.9 | GO:0070695 | FHF complex(GO:0070695) |
| 0.5 | 2.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.5 | 4.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.5 | 2.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.5 | 9.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.5 | 2.5 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.5 | 2.3 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.4 | 19.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.4 | 8.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.4 | 7.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.4 | 3.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.4 | 4.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.4 | 9.0 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.4 | 8.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.4 | 5.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.4 | 10.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.4 | 60.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.4 | 18.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.4 | 4.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.4 | 1.8 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 4.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.3 | 1.4 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.3 | 1.7 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.3 | 1.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.3 | 7.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.3 | 1.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.3 | 6.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.3 | 0.9 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.3 | 6.9 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.3 | 9.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.3 | 1.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.3 | 0.8 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.3 | 2.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 1.6 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.3 | 1.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.3 | 1.3 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.2 | 1.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.2 | 2.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 0.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 0.9 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.2 | 0.7 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.2 | 2.3 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.2 | 3.2 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.2 | 6.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.2 | 0.8 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.2 | 89.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.2 | 4.6 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.2 | 16.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 3.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 2.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 0.9 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 6.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 7.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 2.7 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.2 | 1.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 0.6 | GO:0000802 | transverse filament(GO:0000802) |
| 0.2 | 2.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 3.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.4 | GO:0060473 | cortical granule(GO:0060473) |
| 0.1 | 1.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.7 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 1.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.7 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.7 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 1.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 3.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.0 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 21.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 0.6 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 1.7 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 1.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 2.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.6 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 0.9 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 15.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 4.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 1.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 3.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.9 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 2.1 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 2.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 2.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.3 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 2.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 0.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 8.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 1.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 106.8 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.1 | 0.3 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 2.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 4.8 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.0 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 7.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.9 | GO:0090568 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 1.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0035841 | growing cell tip(GO:0035838) new growing cell tip(GO:0035841) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 3.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.6 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 1.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 11.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 21.8 | 87.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 11.8 | 35.5 | GO:0005186 | pheromone activity(GO:0005186) |
| 10.5 | 62.8 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 8.5 | 25.6 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 6.5 | 19.6 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 5.9 | 71.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 4.8 | 4.8 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 4.4 | 13.1 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 4.2 | 16.7 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 3.7 | 11.2 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 3.7 | 7.4 | GO:0070404 | NADH binding(GO:0070404) |
| 3.5 | 21.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 3.1 | 9.4 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 3.0 | 115.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 3.0 | 178.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 2.9 | 17.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 2.9 | 14.4 | GO:0032810 | sterol response element binding(GO:0032810) |
| 2.8 | 8.5 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 2.8 | 14.0 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 2.7 | 5.4 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 2.6 | 10.3 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 2.5 | 7.6 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 2.5 | 7.4 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 2.4 | 9.5 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 2.3 | 9.4 | GO:0004096 | catalase activity(GO:0004096) |
| 2.3 | 9.3 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 2.3 | 13.9 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 2.2 | 6.7 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 2.2 | 8.8 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 2.1 | 10.7 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 2.0 | 7.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 2.0 | 5.9 | GO:0005118 | sevenless binding(GO:0005118) |
| 1.9 | 26.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 1.9 | 5.6 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.8 | 11.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 1.8 | 11.0 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 1.8 | 16.5 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 1.8 | 5.4 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 1.8 | 8.9 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 1.7 | 5.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 1.6 | 6.6 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 1.6 | 8.2 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 1.5 | 6.0 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 1.5 | 5.9 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.5 | 10.4 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 1.4 | 11.6 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 1.4 | 17.3 | GO:0008430 | selenium binding(GO:0008430) |
| 1.4 | 4.3 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 1.4 | 7.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 1.4 | 4.2 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 1.3 | 6.7 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 1.3 | 5.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 1.3 | 5.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 1.3 | 10.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 1.3 | 3.8 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 1.2 | 3.7 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 1.2 | 19.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 1.2 | 7.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 1.2 | 4.6 | GO:0033814 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 1.1 | 6.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.1 | 22.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 1.1 | 3.3 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 1.1 | 10.8 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 1.0 | 4.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.0 | 3.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 1.0 | 12.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 1.0 | 24.0 | GO:0005550 | pheromone binding(GO:0005550) |
| 1.0 | 4.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.0 | 5.9 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 1.0 | 4.9 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 1.0 | 12.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.9 | 3.8 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.9 | 8.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.9 | 4.7 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.9 | 5.5 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.9 | 6.4 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.9 | 4.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.9 | 2.6 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.9 | 2.6 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.8 | 5.0 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.8 | 7.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.8 | 4.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.8 | 9.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.8 | 5.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.8 | 3.9 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.8 | 2.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.7 | 18.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.7 | 2.8 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.7 | 6.2 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.7 | 3.4 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.7 | 2.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.7 | 6.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.7 | 4.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.7 | 3.9 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.6 | 1.9 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.6 | 7.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.6 | 3.9 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.6 | 1.9 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.6 | 22.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.6 | 3.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.6 | 4.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.6 | 3.7 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.6 | 2.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) efflux transmembrane transporter activity(GO:0015562) |
| 0.6 | 10.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.6 | 9.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.6 | 5.9 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.6 | 3.5 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.6 | 1.7 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.5 | 2.2 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.5 | 2.7 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.5 | 2.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.5 | 2.7 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.5 | 1.6 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.5 | 2.6 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.5 | 3.7 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.5 | 2.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.5 | 1.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.5 | 3.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.5 | 8.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.5 | 3.7 | GO:0035473 | lipase binding(GO:0035473) |
| 0.5 | 1.8 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.5 | 7.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.5 | 0.9 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.5 | 1.4 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.5 | 4.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.4 | 4.4 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.4 | 4.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.4 | 7.9 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.4 | 1.7 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.4 | 1.7 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.4 | 3.0 | GO:0016801 | hydrolase activity, acting on ether bonds(GO:0016801) ether hydrolase activity(GO:0016803) |
| 0.4 | 3.8 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.4 | 8.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.4 | 2.8 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.4 | 2.7 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.4 | 3.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 8.9 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.4 | 1.2 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.4 | 2.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.4 | 4.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.4 | 4.9 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.4 | 13.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.4 | 14.1 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.4 | 3.7 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.4 | 11.6 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.4 | 17.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.3 | 2.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.3 | 3.0 | GO:0046977 | TAP binding(GO:0046977) |
| 0.3 | 19.9 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.3 | 5.0 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.3 | 2.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.3 | 2.6 | GO:0002046 | opsin binding(GO:0002046) |
| 0.3 | 1.3 | GO:0051381 | histamine binding(GO:0051381) |
| 0.3 | 0.6 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.3 | 1.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.3 | 1.2 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.3 | 4.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.3 | 0.8 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.3 | 0.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.3 | 1.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.3 | 6.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 2.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 1.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.3 | 1.0 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) bradykinin receptor binding(GO:0031711) metallodipeptidase activity(GO:0070573) |
| 0.3 | 2.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.3 | 3.3 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.3 | 6.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.3 | 1.8 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 2.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 5.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 1.9 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 2.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 0.9 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.2 | 0.7 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.2 | 5.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 2.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 3.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 0.7 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.2 | 0.9 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.2 | 1.8 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.2 | 1.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 0.9 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 5.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 1.5 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.2 | 2.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 3.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 0.6 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.2 | 3.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 2.9 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 2.4 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.2 | 4.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 1.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 11.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.2 | 1.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 0.7 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.2 | 2.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 2.4 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) |
| 0.2 | 3.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.2 | 0.7 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 1.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 2.9 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 0.8 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.2 | 1.4 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 6.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 0.6 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 4.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.4 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.1 | 5.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.7 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.9 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 3.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 3.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 0.5 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 5.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.4 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 1.6 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.1 | 2.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 1.9 | GO:0043176 | amine binding(GO:0043176) |
| 0.1 | 0.7 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 1.7 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 11.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 7.2 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 0.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 1.6 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 0.5 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 2.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.5 | GO:0038100 | nodal binding(GO:0038100) |
| 0.1 | 6.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.6 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 1.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 7.2 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.1 | 1.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.1 | 0.3 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 15.1 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 4.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 4.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.6 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.3 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 1.4 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.1 | 4.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.0 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 0.7 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 0.5 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 9.8 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.1 | 1.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 3.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 7.8 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 0.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 9.6 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 1.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 1.9 | GO:0016248 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.1 | 1.4 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 2.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.0 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.3 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 1.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 2.1 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 1.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 3.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 5.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 2.1 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 9.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 2.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 1.0 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 4.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.8 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 1.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.0 | GO:0004620 | phospholipase activity(GO:0004620) lipase activity(GO:0016298) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 4.8 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 3.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 20.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.4 | 1.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.3 | 5.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 24.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.3 | 19.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.3 | 16.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.3 | 3.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 4.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.2 | 4.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 5.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 1.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 5.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 1.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 3.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 6.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 2.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 5.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 2.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 1.9 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 0.9 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 0.8 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 3.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 9.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 0.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 1.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 2.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 4.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 4.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 60.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 4.3 | 25.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 3.8 | 98.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 3.6 | 40.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 3.6 | 10.9 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 2.6 | 74.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 2.4 | 36.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 2.1 | 20.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 1.9 | 19.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 1.8 | 23.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 1.5 | 22.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 1.4 | 21.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.3 | 21.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 1.3 | 21.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 1.1 | 19.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 1.1 | 12.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.9 | 12.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.9 | 8.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.8 | 24.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.7 | 10.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.6 | 7.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.6 | 5.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.5 | 3.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.5 | 9.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.5 | 6.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.5 | 9.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.4 | 5.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.4 | 9.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.4 | 10.9 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.4 | 2.7 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.4 | 4.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 4.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.4 | 25.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.4 | 5.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 7.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.3 | 4.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 24.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 4.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.3 | 4.0 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.3 | 2.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 1.8 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.3 | 5.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 26.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 2.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 2.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 4.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 7.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 10.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 2.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.2 | 4.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 6.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 4.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 3.0 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.2 | 8.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 2.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 10.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 2.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 2.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.2 | 7.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 2.0 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.2 | 4.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 2.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 4.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 2.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 7.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 3.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 5.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 2.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 0.7 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.1 | 5.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 4.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 20.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 2.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 1.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 0.8 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.7 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 1.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 2.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.7 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 2.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.0 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 1.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 0.7 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 4.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 5.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.8 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 3.4 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 1.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 2.2 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 2.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.4 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 1.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 1.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.0 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |