Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
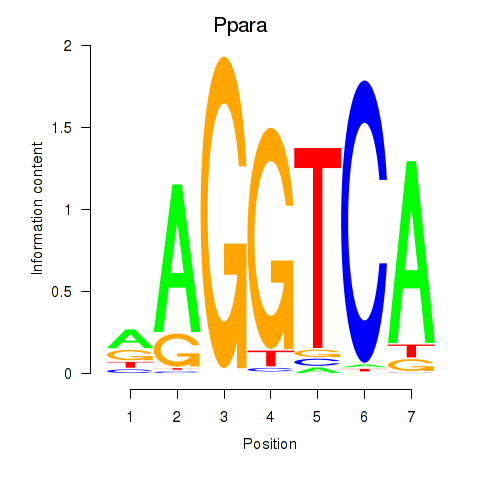
Results for Ppara
Z-value: 0.93
Transcription factors associated with Ppara
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ppara
|
ENSMUSG00000022383.7 | peroxisome proliferator activated receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ppara | mm10_v2_chr15_+_85736107_85736131 | 0.05 | 7.9e-01 | Click! |
Activity profile of Ppara motif
Sorted Z-values of Ppara motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_69617879 | 11.17 |
ENSMUST00000005334.2
|
Shbg
|
sex hormone binding globulin |
| chr7_-_142578093 | 7.55 |
ENSMUST00000149974.1
ENSMUST00000152754.1 |
H19
|
H19 fetal liver mRNA |
| chr7_-_142578139 | 7.50 |
ENSMUST00000136359.1
|
H19
|
H19 fetal liver mRNA |
| chr3_+_114030532 | 5.63 |
ENSMUST00000123619.1
ENSMUST00000092155.5 |
Col11a1
|
collagen, type XI, alpha 1 |
| chr9_+_111019284 | 5.59 |
ENSMUST00000035077.3
|
Ltf
|
lactotransferrin |
| chr8_-_123894736 | 5.46 |
ENSMUST00000034453.4
|
Acta1
|
actin, alpha 1, skeletal muscle |
| chrX_+_101449078 | 4.43 |
ENSMUST00000033674.5
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr12_+_109459843 | 4.26 |
ENSMUST00000173812.1
|
Dlk1
|
delta-like 1 homolog (Drosophila) |
| chr15_+_80091320 | 4.23 |
ENSMUST00000009728.6
ENSMUST00000009727.5 |
Syngr1
|
synaptogyrin 1 |
| chr9_+_65101453 | 4.18 |
ENSMUST00000077696.6
ENSMUST00000035499.4 ENSMUST00000166273.1 |
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr15_-_79285502 | 4.04 |
ENSMUST00000165408.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr11_-_3504766 | 3.95 |
ENSMUST00000044507.5
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chrX_+_157699113 | 3.79 |
ENSMUST00000112521.1
|
Smpx
|
small muscle protein, X-linked |
| chr12_+_109545390 | 3.76 |
ENSMUST00000146701.1
|
Meg3
|
maternally expressed 3 |
| chr6_+_121636173 | 3.76 |
ENSMUST00000032203.7
|
A2m
|
alpha-2-macroglobulin |
| chrX_-_136203637 | 3.50 |
ENSMUST00000151592.1
ENSMUST00000131510.1 ENSMUST00000066819.4 |
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chr16_+_17980565 | 3.45 |
ENSMUST00000075371.3
|
Vpreb2
|
pre-B lymphocyte gene 2 |
| chr2_-_13491900 | 3.40 |
ENSMUST00000091436.5
|
Cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr4_-_119189949 | 3.18 |
ENSMUST00000124626.1
|
Ermap
|
erythroblast membrane-associated protein |
| chr15_-_79285470 | 3.16 |
ENSMUST00000170955.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr12_+_102554966 | 3.13 |
ENSMUST00000021610.5
|
Chga
|
chromogranin A |
| chrX_-_142306170 | 3.11 |
ENSMUST00000134825.2
|
Kcne1l
|
potassium voltage-gated channel, Isk-related family, member 1-like, pseudogene |
| chr16_-_16869255 | 3.11 |
ENSMUST00000075017.4
|
Vpreb1
|
pre-B lymphocyte gene 1 |
| chr2_+_131186942 | 3.10 |
ENSMUST00000028804.8
ENSMUST00000079857.8 |
Cdc25b
|
cell division cycle 25B |
| chr10_-_128401218 | 3.09 |
ENSMUST00000042666.5
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr7_-_100514800 | 2.93 |
ENSMUST00000054923.7
|
Dnajb13
|
DnaJ (Hsp40) related, subfamily B, member 13 |
| chr8_-_104624266 | 2.93 |
ENSMUST00000163783.2
|
Cdh16
|
cadherin 16 |
| chr12_+_109544498 | 2.63 |
ENSMUST00000126289.1
|
Meg3
|
maternally expressed 3 |
| chrX_-_135210672 | 2.62 |
ENSMUST00000033783.1
|
Tceal6
|
transcription elongation factor A (SII)-like 6 |
| chr15_+_35296090 | 2.62 |
ENSMUST00000022952.4
|
Osr2
|
odd-skipped related 2 |
| chr3_+_14886426 | 2.59 |
ENSMUST00000029078.7
|
Car2
|
carbonic anhydrase 2 |
| chr15_-_89425856 | 2.46 |
ENSMUST00000109313.2
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr11_+_69098937 | 2.43 |
ENSMUST00000021271.7
|
Per1
|
period circadian clock 1 |
| chr7_-_110982049 | 2.30 |
ENSMUST00000142368.1
|
Mrvi1
|
MRV integration site 1 |
| chr9_+_51213683 | 2.26 |
ENSMUST00000034554.7
|
Pou2af1
|
POU domain, class 2, associating factor 1 |
| chr17_-_25880236 | 2.26 |
ENSMUST00000176696.1
ENSMUST00000095487.5 |
Wfikkn1
|
WAP, FS, Ig, KU, and NTR-containing protein 1 |
| chr8_+_94977101 | 2.15 |
ENSMUST00000179619.1
|
Gpr56
|
G protein-coupled receptor 56 |
| chr1_-_167393826 | 2.13 |
ENSMUST00000028005.2
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr19_+_8591254 | 2.09 |
ENSMUST00000010251.3
ENSMUST00000170817.1 |
Slc22a8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr11_-_102082464 | 2.04 |
ENSMUST00000100398.4
|
Mpp2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr6_-_67037399 | 2.02 |
ENSMUST00000043098.6
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr14_+_32321987 | 2.01 |
ENSMUST00000022480.7
|
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr7_-_126463100 | 2.00 |
ENSMUST00000032974.6
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr13_-_12340723 | 1.99 |
ENSMUST00000168193.1
ENSMUST00000110616.1 ENSMUST00000064204.7 |
Actn2
|
actinin alpha 2 |
| chr7_-_98162318 | 1.99 |
ENSMUST00000107112.1
|
Capn5
|
calpain 5 |
| chr4_-_132075250 | 1.98 |
ENSMUST00000105970.1
ENSMUST00000105975.1 |
Epb4.1
|
erythrocyte protein band 4.1 |
| chr2_+_155611175 | 1.98 |
ENSMUST00000092995.5
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr4_-_119190005 | 1.97 |
ENSMUST00000138395.1
ENSMUST00000156746.1 |
Ermap
|
erythroblast membrane-associated protein |
| chr17_-_28560704 | 1.96 |
ENSMUST00000114785.1
ENSMUST00000025062.3 |
Clps
|
colipase, pancreatic |
| chr13_+_44729535 | 1.94 |
ENSMUST00000174068.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr6_-_52217505 | 1.94 |
ENSMUST00000048715.6
|
Hoxa7
|
homeobox A7 |
| chr8_+_95055094 | 1.90 |
ENSMUST00000058479.6
|
Ccdc135
|
coiled-coil domain containing 135 |
| chr16_-_20426375 | 1.90 |
ENSMUST00000079158.6
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr1_-_167466780 | 1.86 |
ENSMUST00000036643.4
|
Lrrc52
|
leucine rich repeat containing 52 |
| chr4_-_129491022 | 1.81 |
ENSMUST00000000421.5
|
Tssk3
|
testis-specific serine kinase 3 |
| chrX_-_162964557 | 1.76 |
ENSMUST00000038769.2
|
S100g
|
S100 calcium binding protein G |
| chr9_-_70421533 | 1.73 |
ENSMUST00000034742.6
|
Ccnb2
|
cyclin B2 |
| chr11_-_120551126 | 1.73 |
ENSMUST00000026121.2
|
Ppp1r27
|
protein phosphatase 1, regulatory subunit 27 |
| chr15_-_42676967 | 1.72 |
ENSMUST00000022921.5
|
Angpt1
|
angiopoietin 1 |
| chrX_-_49788204 | 1.72 |
ENSMUST00000114893.1
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr1_+_135783065 | 1.71 |
ENSMUST00000132795.1
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr11_-_70255329 | 1.68 |
ENSMUST00000108574.2
ENSMUST00000000329.2 |
Alox12
|
arachidonate 12-lipoxygenase |
| chr7_-_110982443 | 1.68 |
ENSMUST00000005751.6
|
Mrvi1
|
MRV integration site 1 |
| chr11_-_120648104 | 1.68 |
ENSMUST00000026134.2
|
Myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr7_-_25005895 | 1.68 |
ENSMUST00000102858.3
ENSMUST00000080882.6 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr4_+_34893772 | 1.67 |
ENSMUST00000029975.3
ENSMUST00000135871.1 ENSMUST00000108130.1 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr18_-_56925509 | 1.66 |
ENSMUST00000102912.1
|
March3
|
membrane-associated ring finger (C3HC4) 3 |
| chr4_-_134254076 | 1.66 |
ENSMUST00000060050.5
|
Grrp1
|
glycine/arginine rich protein 1 |
| chr2_+_25242929 | 1.66 |
ENSMUST00000114355.1
ENSMUST00000060818.1 |
Rnf208
|
ring finger protein 208 |
| chr13_+_44729794 | 1.66 |
ENSMUST00000172830.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr15_-_36598019 | 1.66 |
ENSMUST00000155116.1
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr15_-_66801577 | 1.64 |
ENSMUST00000168589.1
|
Sla
|
src-like adaptor |
| chr7_+_110768169 | 1.62 |
ENSMUST00000170374.1
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr9_-_121792478 | 1.61 |
ENSMUST00000035110.4
|
Hhatl
|
hedgehog acyltransferase-like |
| chr15_-_5244178 | 1.60 |
ENSMUST00000047379.8
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr3_+_95164306 | 1.59 |
ENSMUST00000107217.1
ENSMUST00000168321.1 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr2_-_26021679 | 1.59 |
ENSMUST00000036509.7
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr7_+_142441808 | 1.58 |
ENSMUST00000105971.1
ENSMUST00000145287.1 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr5_+_135187251 | 1.58 |
ENSMUST00000002825.5
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr6_+_113531675 | 1.56 |
ENSMUST00000036340.5
ENSMUST00000101051.2 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr15_-_5244164 | 1.56 |
ENSMUST00000120563.1
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr8_-_122460666 | 1.56 |
ENSMUST00000006762.5
|
Snai3
|
snail homolog 3 (Drosophila) |
| chr2_-_131160006 | 1.55 |
ENSMUST00000103188.3
ENSMUST00000133602.1 ENSMUST00000028800.5 |
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene |
| chr5_-_148392810 | 1.55 |
ENSMUST00000138257.1
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr12_-_110978981 | 1.53 |
ENSMUST00000135131.1
ENSMUST00000043459.6 ENSMUST00000128353.1 |
Ankrd9
|
ankyrin repeat domain 9 |
| chr10_-_13324160 | 1.53 |
ENSMUST00000105545.4
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr12_-_40248073 | 1.52 |
ENSMUST00000169926.1
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr7_+_35449035 | 1.52 |
ENSMUST00000118969.1
ENSMUST00000118383.1 |
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr9_-_96437434 | 1.51 |
ENSMUST00000070500.2
|
BC043934
|
cDNA sequence BC043934 |
| chr3_+_68869563 | 1.51 |
ENSMUST00000054551.2
|
1110032F04Rik
|
RIKEN cDNA 1110032F04 gene |
| chr11_-_74590065 | 1.51 |
ENSMUST00000145524.1
ENSMUST00000047488.7 |
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chr7_-_116031047 | 1.51 |
ENSMUST00000106612.1
|
Sox6
|
SRY-box containing gene 6 |
| chr7_-_19715395 | 1.49 |
ENSMUST00000032555.9
ENSMUST00000093552.5 |
Tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr12_+_4082574 | 1.48 |
ENSMUST00000020986.7
|
Dnajc27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr1_+_75400070 | 1.45 |
ENSMUST00000113589.1
|
Speg
|
SPEG complex locus |
| chr11_+_116198853 | 1.45 |
ENSMUST00000021130.6
|
Ten1
|
TEN1 telomerase capping complex subunit |
| chr12_+_4082596 | 1.44 |
ENSMUST00000049584.5
|
Dnajc27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr4_-_117125618 | 1.43 |
ENSMUST00000183310.1
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr4_-_117178726 | 1.43 |
ENSMUST00000153953.1
ENSMUST00000106436.1 |
Kif2c
|
kinesin family member 2C |
| chr15_+_73723131 | 1.43 |
ENSMUST00000165541.1
ENSMUST00000167582.1 |
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr2_-_26021532 | 1.43 |
ENSMUST00000136750.1
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr9_+_96258697 | 1.42 |
ENSMUST00000179416.1
|
Tfdp2
|
transcription factor Dp 2 |
| chrX_-_8145679 | 1.42 |
ENSMUST00000115619.1
ENSMUST00000115617.3 ENSMUST00000040010.3 |
Rbm3
|
RNA binding motif protein 3 |
| chr1_+_86021935 | 1.41 |
ENSMUST00000052854.6
ENSMUST00000125083.1 ENSMUST00000152501.1 ENSMUST00000113344.1 ENSMUST00000130504.1 ENSMUST00000153247.2 |
Spata3
|
spermatogenesis associated 3 |
| chr7_-_110982169 | 1.41 |
ENSMUST00000154466.1
|
Mrvi1
|
MRV integration site 1 |
| chr2_-_29869785 | 1.40 |
ENSMUST00000047607.1
|
2600006K01Rik
|
RIKEN cDNA 2600006K01 gene |
| chr17_-_45474839 | 1.39 |
ENSMUST00000024731.8
|
Spats1
|
spermatogenesis associated, serine-rich 1 |
| chr19_-_4615453 | 1.39 |
ENSMUST00000053597.2
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr12_-_40223149 | 1.38 |
ENSMUST00000171553.1
ENSMUST00000001672.5 |
Ifrd1
|
interferon-related developmental regulator 1 |
| chr15_+_80097866 | 1.38 |
ENSMUST00000143928.1
|
Syngr1
|
synaptogyrin 1 |
| chr12_-_17176888 | 1.37 |
ENSMUST00000170580.1
|
Kcnf1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr11_+_69965396 | 1.37 |
ENSMUST00000018713.6
|
Cldn7
|
claudin 7 |
| chr12_-_110978943 | 1.37 |
ENSMUST00000142012.1
|
Ankrd9
|
ankyrin repeat domain 9 |
| chrX_-_8145713 | 1.37 |
ENSMUST00000115615.2
ENSMUST00000115616.1 ENSMUST00000115621.2 |
Rbm3
|
RNA binding motif protein 3 |
| chr5_-_134946917 | 1.35 |
ENSMUST00000051401.2
|
Cldn4
|
claudin 4 |
| chr7_+_43797567 | 1.33 |
ENSMUST00000085461.2
|
Klk8
|
kallikrein related-peptidase 8 |
| chr7_-_134232125 | 1.33 |
ENSMUST00000127524.1
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr13_+_108214389 | 1.33 |
ENSMUST00000022207.8
|
Elovl7
|
ELOVL family member 7, elongation of long chain fatty acids (yeast) |
| chr3_-_20275659 | 1.32 |
ENSMUST00000011607.5
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr5_+_13399309 | 1.31 |
ENSMUST00000030714.7
ENSMUST00000141968.1 |
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr1_-_58973421 | 1.30 |
ENSMUST00000173590.1
ENSMUST00000027186.5 |
Trak2
|
trafficking protein, kinesin binding 2 |
| chr14_+_55491062 | 1.30 |
ENSMUST00000076236.5
|
Lrrc16b
|
leucine rich repeat containing 16B |
| chr11_+_54304005 | 1.30 |
ENSMUST00000000145.5
ENSMUST00000138515.1 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr10_+_80264942 | 1.28 |
ENSMUST00000105362.1
ENSMUST00000105361.3 |
Dazap1
|
DAZ associated protein 1 |
| chr2_+_112265809 | 1.27 |
ENSMUST00000110991.2
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr3_+_153973436 | 1.27 |
ENSMUST00000089948.5
|
Slc44a5
|
solute carrier family 44, member 5 |
| chrX_-_134583114 | 1.27 |
ENSMUST00000113213.1
ENSMUST00000033617.6 |
Btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chr7_-_4445181 | 1.27 |
ENSMUST00000138798.1
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr19_-_4615647 | 1.26 |
ENSMUST00000113822.2
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr11_+_4236411 | 1.26 |
ENSMUST00000075221.2
|
Osm
|
oncostatin M |
| chr7_+_130865835 | 1.25 |
ENSMUST00000075181.4
ENSMUST00000048180.5 |
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chrX_-_12160355 | 1.25 |
ENSMUST00000043441.6
|
Bcor
|
BCL6 interacting corepressor |
| chr7_+_91090728 | 1.24 |
ENSMUST00000074273.3
|
Dlg2
|
discs, large homolog 2 (Drosophila) |
| chr4_+_156215920 | 1.24 |
ENSMUST00000105572.1
|
2310042D19Rik
|
RIKEN cDNA 2310042D19 gene |
| chr7_+_30751471 | 1.24 |
ENSMUST00000182229.1
ENSMUST00000182227.1 ENSMUST00000080518.6 ENSMUST00000182721.1 |
Sbsn
|
suprabasin |
| chrX_+_136666375 | 1.24 |
ENSMUST00000060904.4
ENSMUST00000113100.1 ENSMUST00000128040.1 |
Tceal3
|
transcription elongation factor A (SII)-like 3 |
| chr1_+_134193432 | 1.24 |
ENSMUST00000038445.6
|
Mybph
|
myosin binding protein H |
| chr2_+_32628390 | 1.24 |
ENSMUST00000156578.1
|
Ak1
|
adenylate kinase 1 |
| chr1_+_75375271 | 1.23 |
ENSMUST00000087122.5
|
Speg
|
SPEG complex locus |
| chr4_+_120588728 | 1.23 |
ENSMUST00000171482.1
|
Gm8439
|
predicted gene 8439 |
| chr16_-_20426322 | 1.22 |
ENSMUST00000115547.2
ENSMUST00000096199.4 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr5_-_18360384 | 1.21 |
ENSMUST00000074694.5
|
Gnai1
|
guanine nucleotide binding protein (G protein), alpha inhibiting 1 |
| chr14_+_54476100 | 1.21 |
ENSMUST00000164766.1
ENSMUST00000164697.1 |
Rem2
|
rad and gem related GTP binding protein 2 |
| chr9_+_65587187 | 1.20 |
ENSMUST00000047099.5
ENSMUST00000131483.1 ENSMUST00000141046.1 |
Pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr2_+_164805082 | 1.20 |
ENSMUST00000052107.4
|
Zswim3
|
zinc finger SWIM-type containing 3 |
| chr7_-_115846080 | 1.19 |
ENSMUST00000166207.1
|
Sox6
|
SRY-box containing gene 6 |
| chr1_-_163289214 | 1.19 |
ENSMUST00000183691.1
|
Prrx1
|
paired related homeobox 1 |
| chr7_+_35449154 | 1.18 |
ENSMUST00000032703.9
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr1_+_12718496 | 1.18 |
ENSMUST00000088585.3
|
Sulf1
|
sulfatase 1 |
| chr17_+_28142267 | 1.17 |
ENSMUST00000043503.3
|
Scube3
|
signal peptide, CUB domain, EGF-like 3 |
| chr14_+_62292475 | 1.17 |
ENSMUST00000166879.1
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr5_+_114003678 | 1.17 |
ENSMUST00000112292.2
|
Dao
|
D-amino acid oxidase |
| chr1_+_135132693 | 1.17 |
ENSMUST00000049449.4
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr5_-_131616599 | 1.16 |
ENSMUST00000161804.1
|
Auts2
|
autism susceptibility candidate 2 |
| chr7_+_24462475 | 1.16 |
ENSMUST00000002284.9
|
Plaur
|
plasminogen activator, urokinase receptor |
| chr4_+_138250403 | 1.16 |
ENSMUST00000105824.1
ENSMUST00000124239.1 ENSMUST00000105818.1 |
Sh2d5
Kif17
|
SH2 domain containing 5 kinesin family member 17 |
| chr15_-_89425795 | 1.13 |
ENSMUST00000168376.1
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr9_+_65587149 | 1.13 |
ENSMUST00000134538.1
ENSMUST00000136205.1 |
Pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr5_+_47984571 | 1.12 |
ENSMUST00000174313.1
|
Slit2
|
slit homolog 2 (Drosophila) |
| chr2_-_163397946 | 1.12 |
ENSMUST00000017961.4
ENSMUST00000109425.2 |
Jph2
|
junctophilin 2 |
| chr1_+_191821444 | 1.12 |
ENSMUST00000027931.7
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2 |
| chr11_+_69964758 | 1.12 |
ENSMUST00000108597.1
ENSMUST00000060651.5 ENSMUST00000108596.1 |
Cldn7
|
claudin 7 |
| chr8_-_4259257 | 1.11 |
ENSMUST00000053252.7
|
Ctxn1
|
cortexin 1 |
| chr11_+_4986824 | 1.11 |
ENSMUST00000009234.9
ENSMUST00000109897.1 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr4_+_138250462 | 1.10 |
ENSMUST00000105823.1
|
Sh2d5
|
SH2 domain containing 5 |
| chr9_-_119578981 | 1.10 |
ENSMUST00000117911.1
ENSMUST00000120420.1 |
Scn5a
|
sodium channel, voltage-gated, type V, alpha |
| chr7_-_4445595 | 1.10 |
ENSMUST00000119485.1
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr4_-_83021102 | 1.09 |
ENSMUST00000071708.5
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr17_-_51826562 | 1.08 |
ENSMUST00000024720.4
ENSMUST00000129667.1 ENSMUST00000156051.1 ENSMUST00000169480.1 ENSMUST00000148559.1 |
Satb1
|
special AT-rich sequence binding protein 1 |
| chr7_+_45335256 | 1.07 |
ENSMUST00000085351.4
|
Hrc
|
histidine rich calcium binding protein |
| chr11_-_40733373 | 1.07 |
ENSMUST00000020579.8
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr3_-_89418287 | 1.07 |
ENSMUST00000029679.3
|
Cks1b
|
CDC28 protein kinase 1b |
| chr17_+_24727819 | 1.07 |
ENSMUST00000170239.2
|
Rpl3l
|
ribosomal protein L3-like |
| chr3_+_122729158 | 1.05 |
ENSMUST00000066728.5
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific |
| chr4_-_41048124 | 1.04 |
ENSMUST00000030136.6
|
Aqp7
|
aquaporin 7 |
| chr3_-_103809278 | 1.04 |
ENSMUST00000063502.6
ENSMUST00000106832.1 ENSMUST00000106834.1 ENSMUST00000029435.8 |
Dclre1b
|
DNA cross-link repair 1B, PSO2 homolog (S. cerevisiae) |
| chr16_-_4523056 | 1.04 |
ENSMUST00000090500.3
ENSMUST00000023161.7 |
Srl
|
sarcalumenin |
| chrX_+_157702574 | 1.03 |
ENSMUST00000112520.1
|
Smpx
|
small muscle protein, X-linked |
| chr14_-_62292959 | 1.03 |
ENSMUST00000063169.8
|
Dleu7
|
deleted in lymphocytic leukemia, 7 |
| chr7_-_128206346 | 1.02 |
ENSMUST00000033049.7
|
Cox6a2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr11_-_61494173 | 1.02 |
ENSMUST00000101085.2
ENSMUST00000079080.6 ENSMUST00000108714.1 |
Mapk7
|
mitogen-activated protein kinase 7 |
| chr6_-_137571007 | 1.02 |
ENSMUST00000100841.2
|
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr6_-_118757974 | 1.01 |
ENSMUST00000112825.2
|
Cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit |
| chrX_-_150589844 | 1.00 |
ENSMUST00000112725.1
ENSMUST00000112720.1 |
Apex2
|
apurinic/apyrimidinic endonuclease 2 |
| chr12_+_85599388 | 1.00 |
ENSMUST00000050687.6
|
Jdp2
|
Jun dimerization protein 2 |
| chr5_+_114003833 | 1.00 |
ENSMUST00000086599.4
|
Dao
|
D-amino acid oxidase |
| chr7_-_142095266 | 0.99 |
ENSMUST00000039926.3
|
Dusp8
|
dual specificity phosphatase 8 |
| chr16_+_84834901 | 0.99 |
ENSMUST00000114184.1
|
Gabpa
|
GA repeat binding protein, alpha |
| chr1_-_170927567 | 0.98 |
ENSMUST00000046322.7
ENSMUST00000159171.1 |
Fcrla
|
Fc receptor-like A |
| chr7_-_134232005 | 0.98 |
ENSMUST00000134504.1
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr1_+_82724881 | 0.98 |
ENSMUST00000078332.6
|
Mff
|
mitochondrial fission factor |
| chr2_-_59160644 | 0.97 |
ENSMUST00000077687.5
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr11_-_96977660 | 0.97 |
ENSMUST00000107626.1
ENSMUST00000107624.1 |
Sp2
|
Sp2 transcription factor |
| chr6_-_136781718 | 0.97 |
ENSMUST00000078095.6
ENSMUST00000032338.7 |
Gucy2c
|
guanylate cyclase 2c |
| chr19_-_46327121 | 0.97 |
ENSMUST00000041391.4
ENSMUST00000096029.5 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr5_-_137741601 | 0.96 |
ENSMUST00000119498.1
ENSMUST00000061789.7 |
Nyap1
|
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
| chr15_-_99457712 | 0.96 |
ENSMUST00000161948.1
|
Nckap5l
|
NCK-associated protein 5-like |
| chr1_-_170927540 | 0.96 |
ENSMUST00000162136.1
ENSMUST00000162887.1 |
Fcrla
|
Fc receptor-like A |
| chr9_+_118506226 | 0.96 |
ENSMUST00000084820.4
|
Golga4
|
golgi autoantigen, golgin subfamily a, 4 |
| chr18_+_77185815 | 0.96 |
ENSMUST00000079618.4
|
St8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ppara
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 1.2 | 15.0 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 1.1 | 3.2 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of integrin activation(GO:0033624) negative regulation of interleukin-1 alpha secretion(GO:0050712) regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) negative regulation of eosinophil migration(GO:2000417) |
| 1.0 | 3.1 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 1.0 | 2.1 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 1.0 | 3.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 1.0 | 2.9 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 1.0 | 2.9 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.9 | 5.6 | GO:1902732 | antifungal humoral response(GO:0019732) positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.9 | 2.7 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.9 | 2.6 | GO:0035672 | positive regulation of cellular pH reduction(GO:0032849) oligopeptide transmembrane transport(GO:0035672) |
| 0.8 | 5.6 | GO:0035989 | tendon development(GO:0035989) |
| 0.7 | 2.2 | GO:0046144 | D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.7 | 2.0 | GO:0090076 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.7 | 2.6 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.6 | 1.7 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.6 | 1.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.5 | 1.6 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.5 | 4.6 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.5 | 2.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.5 | 5.5 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.5 | 2.4 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.4 | 1.3 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.4 | 1.3 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.4 | 1.3 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.4 | 0.4 | GO:1903463 | regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 0.4 | 1.2 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.4 | 1.2 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.4 | 1.2 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.4 | 1.1 | GO:0050924 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.4 | 2.9 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.4 | 1.1 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.4 | 3.2 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.4 | 1.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.3 | 1.0 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.3 | 1.7 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.3 | 3.0 | GO:0033292 | T-tubule organization(GO:0033292) protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.3 | 2.3 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.3 | 1.7 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.3 | 4.6 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.3 | 1.6 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 0.3 | 0.6 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.3 | 8.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.3 | 1.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.3 | 0.9 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.3 | 1.8 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.3 | 1.2 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.3 | 1.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 1.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.3 | 1.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.3 | 0.9 | GO:0060853 | Notch signaling pathway involved in heart induction(GO:0003137) regulation of Notch signaling pathway involved in heart induction(GO:0035480) positive regulation of Notch signaling pathway involved in heart induction(GO:0035481) arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.3 | 0.6 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.3 | 1.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 | 1.7 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.3 | 0.8 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.3 | 1.3 | GO:0021886 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 0.3 | 3.6 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 | 1.5 | GO:0014826 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.3 | 0.8 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.3 | 1.0 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.2 | 2.7 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 2.4 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.2 | 3.4 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.2 | 4.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 0.9 | GO:0015827 | angiotensin-mediated drinking behavior(GO:0003051) aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.2 | 2.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 1.0 | GO:0015793 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.2 | 1.0 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.2 | 1.2 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.2 | 2.5 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.2 | 5.7 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.2 | 0.8 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.2 | 1.7 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.2 | 1.9 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.2 | 1.1 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.2 | 0.7 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 0.7 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.2 | 0.7 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.2 | 0.7 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.2 | 0.9 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.2 | 0.7 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.2 | 3.7 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.2 | 0.2 | GO:0048296 | regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.2 | 0.7 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.2 | 3.6 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.2 | 1.1 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.2 | 1.6 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.2 | 2.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.3 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 0.6 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.1 | 1.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 0.8 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 1.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.8 | GO:1904923 | regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) |
| 0.1 | 2.5 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.8 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 0.7 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 3.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.9 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.7 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.5 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 0.7 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 1.4 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.8 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.1 | 1.0 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 1.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 1.3 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 0.4 | GO:0060574 | bronchiole development(GO:0060435) intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 0.9 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.2 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.1 | 0.5 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.1 | 0.5 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.1 | 0.6 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 1.2 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 | 0.9 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.3 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 1.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.3 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.4 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 1.3 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 4.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.5 | GO:1902564 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.1 | 1.5 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.5 | GO:0034627 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 1.3 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.1 | 0.4 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 1.5 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 0.8 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.3 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 | 0.3 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 | 1.9 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.1 | 0.3 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 1.7 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.4 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 0.3 | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.1 | 0.3 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.7 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 1.5 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 1.3 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.7 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.4 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.6 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.9 | GO:0002551 | mast cell chemotaxis(GO:0002551) |
| 0.1 | 0.7 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 2.8 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.1 | 0.1 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.1 | 0.6 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.5 | GO:1902748 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.8 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.1 | 0.3 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.3 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 0.6 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 1.0 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 0.3 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 0.6 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.1 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.7 | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) |
| 0.1 | 0.2 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 1.2 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.1 | 2.7 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.1 | 0.3 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) positive regulation of sperm motility(GO:1902093) |
| 0.1 | 0.6 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 1.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.6 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.1 | 1.2 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.1 | 1.3 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.5 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 1.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.8 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 0.3 | GO:0061386 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.6 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 1.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.3 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.1 | 0.7 | GO:1901249 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.1 | 0.6 | GO:0090042 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) actin filament reorganization(GO:0090527) |
| 0.1 | 0.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.3 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.1 | GO:1902953 | lysosomal membrane organization(GO:0097212) positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 | 0.6 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 | 0.2 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 0.6 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 2.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.2 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.2 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.8 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.1 | 0.3 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 | 1.0 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.1 | 0.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 6.3 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.1 | 1.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.3 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 0.7 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.3 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 1.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.8 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.2 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) activation of meiosis involved in egg activation(GO:0060466) |
| 0.1 | 0.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 1.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.3 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 0.3 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.4 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 1.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.6 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.5 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.5 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 2.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 1.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 | 0.2 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 2.8 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.2 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.0 | 1.6 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.3 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.2 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 3.1 | GO:0008361 | regulation of cell size(GO:0008361) |
| 0.0 | 0.4 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.7 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.8 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.2 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.1 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.0 | 0.9 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 9.4 | GO:0048588 | developmental cell growth(GO:0048588) |
| 0.0 | 0.5 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.0 | 0.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.7 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.1 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 | 0.2 | GO:1903012 | positive regulation of bone development(GO:1903012) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 1.3 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0021623 | oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.0 | 0.1 | GO:0009814 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.7 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 1.3 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 1.9 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 1.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.5 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 | 0.3 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.4 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 0.3 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.1 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.0 | 0.7 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.6 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 1.7 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.1 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 1.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.8 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.4 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.4 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.4 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.6 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.9 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 1.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.4 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.2 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.3 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.0 | 0.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.1 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 1.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.8 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.1 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 2.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.7 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.4 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:1903525 | regulation of membrane tubulation(GO:1903525) positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.2 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.0 | 0.1 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.1 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.0 | 0.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.5 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.2 | GO:0045823 | positive regulation of heart contraction(GO:0045823) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.0 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.0 | 0.3 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 2.8 | GO:0007409 | axonogenesis(GO:0007409) |
| 0.0 | 0.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.6 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.7 | 4.8 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.7 | 3.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.6 | 3.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.6 | 7.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.5 | 0.5 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.4 | 5.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 0.8 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.4 | 1.6 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.3 | 2.0 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 0.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.3 | 2.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.2 | 2.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 16.4 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 2.0 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.2 | 1.2 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.2 | 9.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 0.9 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 1.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 0.9 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.2 | 0.5 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 0.8 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.2 | 0.8 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.2 | 1.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 1.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 0.5 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 3.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.7 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 1.4 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 1.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 3.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.0 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.3 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 0.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.4 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 1.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 1.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.4 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 1.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 3.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 6.3 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 2.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 0.5 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 1.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 5.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 2.1 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.2 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 0.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 4.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 1.3 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 4.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.8 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 1.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.2 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.5 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.8 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 1.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 1.3 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.6 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 1.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 3.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.6 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 5.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 5.0 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 6.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 2.1 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 2.7 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 3.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.7 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.3 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 4.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.6 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 1.3 | 3.8 | GO:0019959 | interleukin-8 binding(GO:0019959) tumor necrosis factor binding(GO:0043120) |
| 0.9 | 2.7 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.8 | 2.3 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 0.7 | 3.6 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.5 | 2.2 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.5 | 3.5 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.5 | 2.9 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.4 | 1.3 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.4 | 1.7 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.4 | 2.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.4 | 1.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.4 | 1.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.3 | 1.3 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.3 | 0.9 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.3 | 3.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.3 | 2.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.3 | 2.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 2.4 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 1.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.3 | 2.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.3 | 0.8 | GO:0004771 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 0.3 | 1.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 1.8 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 2.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 1.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 0.7 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 0.7 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.2 | 2.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 1.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 1.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 2.1 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.2 | 1.0 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.2 | 1.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 0.6 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.2 | 0.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 3.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 1.5 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.2 | 0.6 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 1.7 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 0.6 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.2 | 1.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 0.7 | GO:0038100 | nodal binding(GO:0038100) |
| 0.2 | 0.7 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.2 | 0.5 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.2 | 1.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 1.0 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.2 | 0.5 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.2 | 0.8 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 2.0 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.2 | 5.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 1.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 0.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 1.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.2 | 1.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 2.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 0.8 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 1.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.4 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 0.8 | GO:0002135 | CTP binding(GO:0002135) sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.9 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 1.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 5.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.8 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 1.0 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 2.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.7 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 1.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 1.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.9 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.5 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 2.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.7 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 1.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 4.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 0.3 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.4 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 1.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 1.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.3 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 0.3 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.6 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 2.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.3 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 1.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 1.9 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 3.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.4 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 2.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.6 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 3.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 3.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.9 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 1.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.7 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 1.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.2 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 1.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 2.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 2.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.5 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.0 | 2.7 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 6.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 4.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 1.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0002134 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.8 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.3 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 5.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.9 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 1.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.4 | GO:0016722 | oxidoreductase activity, oxidizing metal ions(GO:0016722) |
| 0.0 | 2.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.6 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 4.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.5 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 1.0 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.3 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.1 | GO:0008941 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.0 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 7.6 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.0 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 12.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 5.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 5.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 6.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 5.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 8.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 3.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 1.5 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 2.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 1.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 3.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.8 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.6 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 1.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.8 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.3 | 4.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 1.7 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 6.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 2.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 5.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 2.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 2.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 4.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 2.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 2.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 4.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 2.8 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 2.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.8 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 2.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 6.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.7 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 1.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 2.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 3.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 0.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 3.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.1 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.1 | 1.4 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.1 | 0.9 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 2.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 2.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 2.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 1.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.9 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.7 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 1.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.4 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.1 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.7 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |