Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Pou3f2
Z-value: 0.85
Transcription factors associated with Pou3f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou3f2
|
ENSMUSG00000095139.1 | POU domain, class 3, transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f2 | mm10_v2_chr4_-_22488296_22488366 | 0.28 | 9.6e-02 | Click! |
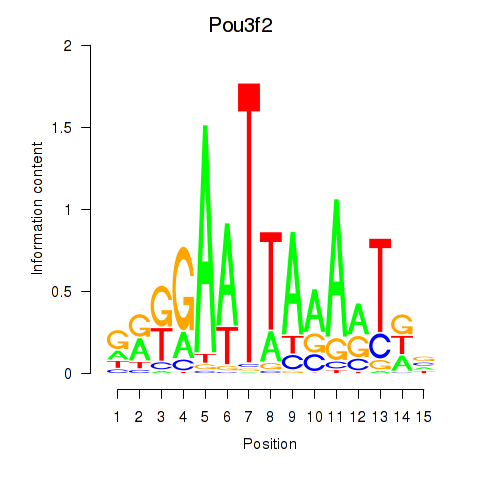
Activity profile of Pou3f2 motif
Sorted Z-values of Pou3f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_41302265 | 5.78 |
ENSMUST00000031913.4
|
Try4
|
trypsin 4 |
| chr2_-_62483637 | 4.04 |
ENSMUST00000136686.1
ENSMUST00000102733.3 |
Gcg
|
glucagon |
| chr7_-_142661858 | 3.57 |
ENSMUST00000145896.2
|
Igf2
|
insulin-like growth factor 2 |
| chr6_+_41354105 | 3.11 |
ENSMUST00000072103.5
|
Try10
|
trypsin 10 |
| chr8_+_21734490 | 2.45 |
ENSMUST00000080533.5
|
Defa24
|
defensin, alpha, 24 |
| chr8_+_21391811 | 2.42 |
ENSMUST00000120874.3
|
Gm21002
|
predicted gene, 21002 |
| chr7_+_13623967 | 2.21 |
ENSMUST00000108525.2
|
Sult2a5
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 5 |
| chr8_+_21529031 | 2.15 |
ENSMUST00000084041.3
|
Gm15308
|
predicted gene 15308 |
| chr6_+_142298419 | 2.09 |
ENSMUST00000041993.2
|
Iapp
|
islet amyloid polypeptide |
| chr14_+_80000292 | 1.97 |
ENSMUST00000088735.3
|
Olfm4
|
olfactomedin 4 |
| chr8_+_21509258 | 1.78 |
ENSMUST00000084042.3
|
Defa20
|
defensin, alpha, 20 |
| chr8_+_21378560 | 1.57 |
ENSMUST00000170275.2
|
Defa2
|
defensin, alpha, 2 |
| chrX_-_139085230 | 1.49 |
ENSMUST00000152457.1
|
Serpina7
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr18_+_5593566 | 1.48 |
ENSMUST00000160910.1
|
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr6_+_40964760 | 1.35 |
ENSMUST00000076638.5
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chrX_-_145505175 | 1.25 |
ENSMUST00000143610.1
|
Amot
|
angiomotin |
| chr5_-_148392810 | 1.21 |
ENSMUST00000138257.1
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chrX_-_139085211 | 1.20 |
ENSMUST00000033626.8
ENSMUST00000060824.3 |
Serpina7
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr7_+_75610038 | 1.14 |
ENSMUST00000125771.1
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr3_+_88965812 | 1.09 |
ENSMUST00000090933.4
|
Ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr14_+_84443553 | 1.06 |
ENSMUST00000071370.5
|
Pcdh17
|
protocadherin 17 |
| chr12_-_91384403 | 0.97 |
ENSMUST00000141429.1
|
Cep128
|
centrosomal protein 128 |
| chr17_+_43389436 | 0.96 |
ENSMUST00000113599.1
|
Gpr116
|
G protein-coupled receptor 116 |
| chrX_-_95166307 | 0.96 |
ENSMUST00000113873.2
ENSMUST00000113876.2 ENSMUST00000113885.1 ENSMUST00000113883.1 ENSMUST00000182001.1 ENSMUST00000113882.1 ENSMUST00000113878.1 ENSMUST00000182562.1 |
Arhgef9
|
CDC42 guanine nucleotide exchange factor (GEF) 9 |
| chrX_+_96456362 | 0.95 |
ENSMUST00000079322.5
ENSMUST00000113838.1 |
Heph
|
hephaestin |
| chr14_-_103844173 | 0.94 |
ENSMUST00000022718.3
|
Ednrb
|
endothelin receptor type B |
| chr14_+_99046406 | 0.92 |
ENSMUST00000022656.6
|
Bora
|
bora, aurora kinase A activator |
| chr14_-_47418407 | 0.91 |
ENSMUST00000043296.3
|
Dlgap5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr14_+_32166104 | 0.84 |
ENSMUST00000164341.1
|
Ncoa4
|
nuclear receptor coactivator 4 |
| chr14_-_103843685 | 0.81 |
ENSMUST00000172237.1
|
Ednrb
|
endothelin receptor type B |
| chr6_-_129740484 | 0.77 |
ENSMUST00000050385.5
|
Klri2
|
killer cell lectin-like receptor family I member 2 |
| chr2_-_62573813 | 0.73 |
ENSMUST00000174234.1
ENSMUST00000000402.9 ENSMUST00000174448.1 |
Fap
|
fibroblast activation protein |
| chr3_-_36571952 | 0.71 |
ENSMUST00000029270.3
|
Ccna2
|
cyclin A2 |
| chr3_-_30013388 | 0.71 |
ENSMUST00000108270.3
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr13_-_21468474 | 0.70 |
ENSMUST00000068235.4
|
Nkapl
|
NFKB activating protein-like |
| chr10_-_68541842 | 0.70 |
ENSMUST00000020103.2
|
1700040L02Rik
|
RIKEN cDNA 1700040L02 gene |
| chr13_+_44840686 | 0.68 |
ENSMUST00000173906.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr19_+_38930909 | 0.68 |
ENSMUST00000025965.5
|
Hells
|
helicase, lymphoid specific |
| chr1_+_34121250 | 0.66 |
ENSMUST00000183006.1
|
Dst
|
dystonin |
| chr3_+_84666192 | 0.65 |
ENSMUST00000107682.1
|
Tmem154
|
transmembrane protein 154 |
| chr11_+_11487671 | 0.65 |
ENSMUST00000020410.4
|
4930415F15Rik
|
RIKEN cDNA 4930415F15 gene |
| chr8_+_21249713 | 0.64 |
ENSMUST00000098895.2
|
Gm15292
|
predicted gene 15292 |
| chr18_+_37400845 | 0.63 |
ENSMUST00000057228.1
|
Pcdhb9
|
protocadherin beta 9 |
| chr5_+_107497762 | 0.63 |
ENSMUST00000152474.1
ENSMUST00000060553.7 |
A830010M20Rik
|
RIKEN cDNA A830010M20 gene |
| chrX_-_165004829 | 0.63 |
ENSMUST00000114890.2
|
Gm17604
|
predicted gene, 17604 |
| chr13_-_74376566 | 0.62 |
ENSMUST00000091481.2
|
Zfp72
|
zinc finger protein 72 |
| chr1_+_74661794 | 0.62 |
ENSMUST00000129890.1
|
Ttll4
|
tubulin tyrosine ligase-like family, member 4 |
| chr1_+_150392794 | 0.61 |
ENSMUST00000124973.2
|
Tpr
|
translocated promoter region |
| chr7_+_16781341 | 0.59 |
ENSMUST00000108496.2
|
Slc1a5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr6_-_129913166 | 0.59 |
ENSMUST00000169901.2
ENSMUST00000167079.2 ENSMUST00000014683.6 |
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chr5_+_86071734 | 0.57 |
ENSMUST00000031171.7
|
Stap1
|
signal transducing adaptor family member 1 |
| chr12_+_69241832 | 0.57 |
ENSMUST00000063445.6
|
Klhdc1
|
kelch domain containing 1 |
| chr11_-_99244058 | 0.56 |
ENSMUST00000103132.3
ENSMUST00000038214.6 |
Krt222
|
keratin 222 |
| chr18_+_4920509 | 0.52 |
ENSMUST00000126977.1
|
Svil
|
supervillin |
| chr10_+_97479470 | 0.51 |
ENSMUST00000105287.3
|
Dcn
|
decorin |
| chr16_-_36784784 | 0.50 |
ENSMUST00000165531.1
|
Slc15a2
|
solute carrier family 15 (H+/peptide transporter), member 2 |
| chr11_+_23256001 | 0.50 |
ENSMUST00000020538.6
ENSMUST00000109551.1 ENSMUST00000102870.1 ENSMUST00000102869.1 |
Xpo1
|
exportin 1, CRM1 homolog (yeast) |
| chr16_-_76022266 | 0.50 |
ENSMUST00000114240.1
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr2_-_136387929 | 0.50 |
ENSMUST00000035264.2
ENSMUST00000077200.3 |
Pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chrX_-_145505136 | 0.49 |
ENSMUST00000112835.1
|
Amot
|
angiomotin |
| chr7_+_30699783 | 0.49 |
ENSMUST00000013227.7
|
2200002J24Rik
|
RIKEN cDNA 2200002J24 gene |
| chr13_+_25056206 | 0.49 |
ENSMUST00000069614.6
|
Dcdc2a
|
doublecortin domain containing 2a |
| chr12_+_51377560 | 0.49 |
ENSMUST00000021335.5
|
Scfd1
|
Sec1 family domain containing 1 |
| chr1_-_52490736 | 0.48 |
ENSMUST00000170269.1
|
Nab1
|
Ngfi-A binding protein 1 |
| chr7_-_128206346 | 0.47 |
ENSMUST00000033049.7
|
Cox6a2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr1_+_161494649 | 0.45 |
ENSMUST00000086084.1
|
Tnfsf18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr5_+_14025305 | 0.44 |
ENSMUST00000073957.6
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr19_-_4877882 | 0.44 |
ENSMUST00000006626.3
|
Actn3
|
actinin alpha 3 |
| chr15_-_83149270 | 0.43 |
ENSMUST00000129372.1
|
Poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr16_+_44811733 | 0.42 |
ENSMUST00000176819.1
ENSMUST00000176321.1 |
Cd200r4
|
CD200 receptor 4 |
| chr14_+_73138755 | 0.42 |
ENSMUST00000171070.1
|
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chrX_+_164419782 | 0.42 |
ENSMUST00000033754.7
|
Piga
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr15_-_75238768 | 0.41 |
ENSMUST00000089689.3
|
Gm10238
|
predicted pseudogene 10238 |
| chr8_+_21618183 | 0.40 |
ENSMUST00000074343.5
|
Defa26
|
defensin, alpha, 26 |
| chr5_-_66054499 | 0.38 |
ENSMUST00000145625.1
|
Rbm47
|
RNA binding motif protein 47 |
| chr2_-_60125651 | 0.37 |
ENSMUST00000112550.1
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr12_-_31351368 | 0.37 |
ENSMUST00000110857.3
|
Dld
|
dihydrolipoamide dehydrogenase |
| chr14_+_79515618 | 0.35 |
ENSMUST00000110835.1
|
Elf1
|
E74-like factor 1 |
| chr6_-_30958990 | 0.35 |
ENSMUST00000101589.3
|
Klf14
|
Kruppel-like factor 14 |
| chr16_-_36784924 | 0.35 |
ENSMUST00000168279.1
ENSMUST00000164579.1 ENSMUST00000023616.2 |
Slc15a2
|
solute carrier family 15 (H+/peptide transporter), member 2 |
| chr1_+_74661714 | 0.33 |
ENSMUST00000042125.8
ENSMUST00000141119.1 |
Ttll4
|
tubulin tyrosine ligase-like family, member 4 |
| chr13_+_108046411 | 0.33 |
ENSMUST00000095458.4
|
Smim15
|
small integral membrane protein 15 |
| chr16_+_36875119 | 0.33 |
ENSMUST00000135406.1
ENSMUST00000114812.1 ENSMUST00000134616.1 ENSMUST00000023534.6 |
Golgb1
Golgb1
|
golgi autoantigen, golgin subfamily b, macrogolgin 1 golgi autoantigen, golgin subfamily b, macrogolgin 1 |
| chr3_+_27984145 | 0.33 |
ENSMUST00000067757.4
|
Pld1
|
phospholipase D1 |
| chr5_+_86804508 | 0.32 |
ENSMUST00000038384.7
|
Ythdc1
|
YTH domain containing 1 |
| chr2_+_104590453 | 0.31 |
ENSMUST00000028599.7
|
Cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr7_-_133015248 | 0.31 |
ENSMUST00000169570.1
|
Ctbp2
|
C-terminal binding protein 2 |
| chr15_-_63997969 | 0.31 |
ENSMUST00000164532.1
|
Fam49b
|
family with sequence similarity 49, member B |
| chr4_-_149126688 | 0.31 |
ENSMUST00000030815.2
|
Cort
|
cortistatin |
| chr2_+_61593077 | 0.31 |
ENSMUST00000112495.1
ENSMUST00000112501.2 |
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr3_+_95929325 | 0.30 |
ENSMUST00000171368.1
ENSMUST00000168106.1 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr17_-_46032366 | 0.30 |
ENSMUST00000071648.5
ENSMUST00000142351.2 ENSMUST00000167860.1 |
Vegfa
|
vascular endothelial growth factor A |
| chr6_+_8209216 | 0.30 |
ENSMUST00000040017.7
|
Mios
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr2_+_80315461 | 0.30 |
ENSMUST00000028392.7
|
Dnajc10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr14_-_88471396 | 0.30 |
ENSMUST00000061628.5
|
Pcdh20
|
protocadherin 20 |
| chr9_+_88581036 | 0.27 |
ENSMUST00000164661.2
|
Trim43a
|
tripartite motif-containing 43A |
| chr4_+_11758147 | 0.27 |
ENSMUST00000029871.5
ENSMUST00000108303.1 |
Cdh17
|
cadherin 17 |
| chr14_-_73049107 | 0.27 |
ENSMUST00000044664.4
ENSMUST00000169168.1 |
Cysltr2
|
cysteinyl leukotriene receptor 2 |
| chr3_+_95929246 | 0.26 |
ENSMUST00000165307.1
ENSMUST00000015893.6 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr13_+_96388294 | 0.26 |
ENSMUST00000099295.4
|
Poc5
|
POC5 centriolar protein homolog (Chlamydomonas) |
| chr2_-_62573905 | 0.26 |
ENSMUST00000102732.3
|
Fap
|
fibroblast activation protein |
| chr17_-_6449571 | 0.26 |
ENSMUST00000180035.1
|
Tmem181b-ps
|
transmembrane protein 181B, pseudogene |
| chr6_+_24748325 | 0.26 |
ENSMUST00000031691.2
|
Hyal4
|
hyaluronoglucosaminidase 4 |
| chr3_+_66985647 | 0.26 |
ENSMUST00000162362.1
ENSMUST00000065074.7 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr8_+_21315546 | 0.25 |
ENSMUST00000168340.1
ENSMUST00000167683.2 |
Gm15299
|
predicted pseudogene 15299 |
| chr5_+_107497718 | 0.25 |
ENSMUST00000112671.2
|
A830010M20Rik
|
RIKEN cDNA A830010M20 gene |
| chr4_-_108780503 | 0.24 |
ENSMUST00000106658.1
|
Zfyve9
|
zinc finger, FYVE domain containing 9 |
| chr6_-_99632376 | 0.23 |
ENSMUST00000176255.1
|
Gm20696
|
predicted gene 20696 |
| chr10_-_53630439 | 0.22 |
ENSMUST00000075540.5
|
Mcm9
|
minichromosome maintenance complex component 9 |
| chr1_-_150392719 | 0.21 |
ENSMUST00000006167.6
ENSMUST00000094477.2 ENSMUST00000097547.3 |
BC003331
|
cDNA sequence BC003331 |
| chr3_+_66985947 | 0.20 |
ENSMUST00000161726.1
ENSMUST00000160504.1 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr11_-_50931612 | 0.20 |
ENSMUST00000109124.3
|
Zfp354b
|
zinc finger protein 354B |
| chr3_+_66985680 | 0.20 |
ENSMUST00000065047.6
|
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr12_+_71170589 | 0.20 |
ENSMUST00000129376.1
|
2700049A03Rik
|
RIKEN cDNA 2700049A03 gene |
| chr4_-_108780782 | 0.19 |
ENSMUST00000106657.1
|
Zfyve9
|
zinc finger, FYVE domain containing 9 |
| chr19_+_60144682 | 0.19 |
ENSMUST00000065383.4
|
E330013P04Rik
|
RIKEN cDNA E330013P04 gene |
| chr18_-_43477764 | 0.19 |
ENSMUST00000057110.9
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr3_+_66985700 | 0.18 |
ENSMUST00000046542.6
ENSMUST00000162693.1 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr11_+_23256566 | 0.18 |
ENSMUST00000136235.1
|
Xpo1
|
exportin 1, CRM1 homolog (yeast) |
| chr7_+_104218787 | 0.17 |
ENSMUST00000098180.3
|
Trim6
|
tripartite motif-containing 6 |
| chr3_-_51560816 | 0.14 |
ENSMUST00000037141.7
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr11_+_70023905 | 0.14 |
ENSMUST00000124568.2
|
Dlg4
|
discs, large homolog 4 (Drosophila) |
| chr15_+_36179530 | 0.14 |
ENSMUST00000171205.1
|
Spag1
|
sperm associated antigen 1 |
| chr10_+_56377300 | 0.13 |
ENSMUST00000068581.7
|
Gja1
|
gap junction protein, alpha 1 |
| chr9_+_88839164 | 0.12 |
ENSMUST00000163255.2
|
Trim43c
|
tripartite motif-containing 43C |
| chr19_-_20390944 | 0.12 |
ENSMUST00000025561.7
|
Anxa1
|
annexin A1 |
| chr6_+_34384218 | 0.12 |
ENSMUST00000038383.7
ENSMUST00000115051.1 |
Akr1b10
|
aldo-keto reductase family 1, member B10 (aldose reductase) |
| chr8_+_83608175 | 0.11 |
ENSMUST00000005620.8
|
Dnajb1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr6_+_30512283 | 0.11 |
ENSMUST00000031798.7
|
Ssmem1
|
serine-rich single-pass membrane protein 1 |
| chr5_+_122284365 | 0.11 |
ENSMUST00000053426.8
|
Pptc7
|
PTC7 protein phosphatase homolog (S. cerevisiae) |
| chr2_-_79908428 | 0.09 |
ENSMUST00000102652.3
ENSMUST00000102651.3 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr1_-_162813926 | 0.08 |
ENSMUST00000144916.1
ENSMUST00000140274.1 |
Fmo4
|
flavin containing monooxygenase 4 |
| chr9_-_105521147 | 0.08 |
ENSMUST00000176770.1
ENSMUST00000085133.6 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr1_-_9298499 | 0.06 |
ENSMUST00000132064.1
|
Sntg1
|
syntrophin, gamma 1 |
| chr7_+_28277706 | 0.06 |
ENSMUST00000094651.2
|
Eid2b
|
EP300 interacting inhibitor of differentiation 2B |
| chr19_-_6543019 | 0.03 |
ENSMUST00000113451.1
|
Slc22a12
|
solute carrier family 22 (organic anion/cation transporter), member 12 |
| chr11_+_72689997 | 0.02 |
ENSMUST00000155998.1
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr8_-_41041828 | 0.02 |
ENSMUST00000051379.7
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr2_-_79908389 | 0.02 |
ENSMUST00000090756.4
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr10_-_69352886 | 0.02 |
ENSMUST00000119827.1
ENSMUST00000020099.5 |
Cdk1
|
cyclin-dependent kinase 1 |
| chr2_-_84650676 | 0.01 |
ENSMUST00000067232.3
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr3_+_141465592 | 0.01 |
ENSMUST00000130636.1
|
Unc5c
|
unc-5 homolog C (C. elegans) |
| chr3_+_55461758 | 0.00 |
ENSMUST00000070418.4
|
Dclk1
|
doublecortin-like kinase 1 |
| chr2_+_178193075 | 0.00 |
ENSMUST00000103065.1
|
Phactr3
|
phosphatase and actin regulator 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou3f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.6 | 4.0 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.4 | 3.6 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.4 | 1.7 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.3 | 1.0 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.3 | 1.7 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.3 | 2.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.2 | 0.6 | GO:0046832 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.2 | 0.6 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.2 | 0.9 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.6 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 0.4 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.1 | 0.4 | GO:1904023 | regulation of fermentation(GO:0043465) regulation of bone mineralization involved in bone maturation(GO:1900157) regulation of NAD metabolic process(GO:1902688) regulation of glucose catabolic process to lactate via pyruvate(GO:1904023) |
| 0.1 | 1.1 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 1.5 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 0.4 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.1 | 4.2 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.3 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 0.7 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 1.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.1 | 0.3 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.9 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.6 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 1.2 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 8.7 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.3 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.7 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0038190 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.0 | 0.7 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.6 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.5 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 2.0 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.7 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.9 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 1.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0032468 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.4 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.8 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.3 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.7 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 1.1 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.5 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.4 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.0 | 0.1 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.1 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 2.0 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.4 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.7 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.6 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.7 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.9 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.8 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 1.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.4 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 2.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 21.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.2 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 1.7 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.3 | 2.2 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.3 | 0.9 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.2 | 0.6 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.2 | 1.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 1.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.3 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 1.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 0.9 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.1 | 0.9 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 3.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.4 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 6.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 9.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.3 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 2.7 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 1.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.3 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.7 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 3.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 2.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 2.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 3.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |