Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Pbx2
Z-value: 1.12
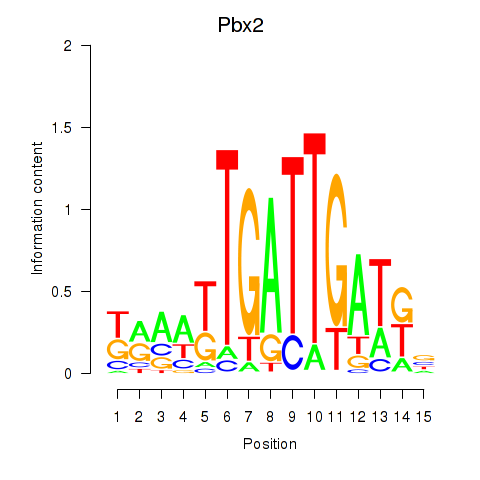
Transcription factors associated with Pbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pbx2
|
ENSMUSG00000034673.8 | pre B cell leukemia homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pbx2 | mm10_v2_chr17_+_34592248_34592329 | -0.91 | 6.4e-15 | Click! |
Activity profile of Pbx2 motif
Sorted Z-values of Pbx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_62054112 | 13.23 |
ENSMUST00000074018.3
|
Mup20
|
major urinary protein 20 |
| chr13_-_23914998 | 11.86 |
ENSMUST00000021769.8
ENSMUST00000110407.2 |
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr3_+_138415484 | 10.88 |
ENSMUST00000161312.1
ENSMUST00000013458.8 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr7_-_46742979 | 9.20 |
ENSMUST00000128088.1
|
Saa1
|
serum amyloid A 1 |
| chr7_+_46751832 | 8.86 |
ENSMUST00000075982.2
|
Saa2
|
serum amyloid A 2 |
| chrX_-_8193387 | 6.52 |
ENSMUST00000143223.1
ENSMUST00000033509.8 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chr11_-_58613481 | 5.54 |
ENSMUST00000048801.7
|
2210407C18Rik
|
RIKEN cDNA 2210407C18 gene |
| chr12_+_108334341 | 5.47 |
ENSMUST00000021684.4
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr3_-_107943390 | 5.14 |
ENSMUST00000106681.1
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr3_-_107943705 | 5.07 |
ENSMUST00000106680.1
ENSMUST00000106684.1 ENSMUST00000106685.2 |
Gstm6
|
glutathione S-transferase, mu 6 |
| chr5_-_87424201 | 5.02 |
ENSMUST00000072818.5
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr4_-_96664112 | 4.86 |
ENSMUST00000030299.7
|
Cyp2j5
|
cytochrome P450, family 2, subfamily j, polypeptide 5 |
| chr3_-_107943362 | 4.45 |
ENSMUST00000106683.1
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr18_-_31820413 | 4.11 |
ENSMUST00000165131.2
|
Gm6665
|
predicted gene 6665 |
| chr11_+_70213910 | 4.07 |
ENSMUST00000171032.1
|
Slc16a11
|
solute carrier family 16 (monocarboxylic acid transporters), member 11 |
| chr18_+_20247340 | 3.84 |
ENSMUST00000054128.6
|
Dsg1c
|
desmoglein 1 gamma |
| chr8_-_94696223 | 3.83 |
ENSMUST00000034227.4
|
Pllp
|
plasma membrane proteolipid |
| chr6_-_127109517 | 3.67 |
ENSMUST00000039913.8
|
9630033F20Rik
|
RIKEN cDNA 9630033F20 gene |
| chr2_-_62412219 | 3.56 |
ENSMUST00000047812.7
|
Dpp4
|
dipeptidylpeptidase 4 |
| chr7_+_51880312 | 3.56 |
ENSMUST00000145049.1
|
Gas2
|
growth arrest specific 2 |
| chr1_+_88138364 | 3.52 |
ENSMUST00000014263.4
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr4_+_141242850 | 3.34 |
ENSMUST00000138096.1
ENSMUST00000006618.2 ENSMUST00000125392.1 |
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr14_+_65969714 | 3.18 |
ENSMUST00000153460.1
|
Clu
|
clusterin |
| chr19_+_39992424 | 3.12 |
ENSMUST00000049178.2
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr3_+_130617645 | 2.94 |
ENSMUST00000163620.1
|
Etnppl
|
ethanolamine phosphate phospholyase |
| chr11_+_97685794 | 2.84 |
ENSMUST00000107584.1
ENSMUST00000107585.2 |
Cisd3
|
CDGSH iron sulfur domain 3 |
| chr5_-_87140318 | 2.83 |
ENSMUST00000067790.6
ENSMUST00000113327.1 |
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr11_-_106580594 | 2.69 |
ENSMUST00000153870.1
|
Tex2
|
testis expressed gene 2 |
| chr8_-_110039330 | 2.61 |
ENSMUST00000109222.2
|
Chst4
|
carbohydrate (chondroitin 6/keratan) sulfotransferase 4 |
| chr6_+_8259288 | 2.59 |
ENSMUST00000159335.1
|
Gm16039
|
predicted gene 16039 |
| chr2_-_58052832 | 2.58 |
ENSMUST00000090940.5
|
Ermn
|
ermin, ERM-like protein |
| chr17_+_37193889 | 2.44 |
ENSMUST00000038844.6
|
Ubd
|
ubiquitin D |
| chr2_-_35061431 | 2.39 |
ENSMUST00000028233.3
|
Hc
|
hemolytic complement |
| chr13_+_4049001 | 2.38 |
ENSMUST00000118717.2
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr3_+_121953213 | 2.36 |
ENSMUST00000037958.7
ENSMUST00000128366.1 |
Arhgap29
|
Rho GTPase activating protein 29 |
| chr11_-_98149551 | 2.34 |
ENSMUST00000103143.3
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr3_-_89393629 | 2.33 |
ENSMUST00000124783.1
ENSMUST00000126027.1 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr2_+_177897096 | 2.32 |
ENSMUST00000108935.1
|
Gm14327
|
predicted gene 14327 |
| chr2_+_27676440 | 2.31 |
ENSMUST00000129514.1
|
Rxra
|
retinoid X receptor alpha |
| chr2_-_86347764 | 2.31 |
ENSMUST00000099894.2
|
Olfr1055
|
olfactory receptor 1055 |
| chr9_+_73101836 | 2.24 |
ENSMUST00000172578.1
|
Khdc3
|
KH domain containing 3, subcortical maternal complex member |
| chr11_+_97685903 | 2.13 |
ENSMUST00000107583.2
|
Cisd3
|
CDGSH iron sulfur domain 3 |
| chr6_+_8259379 | 2.11 |
ENSMUST00000162034.1
ENSMUST00000160705.1 ENSMUST00000159433.1 |
Gm16039
|
predicted gene 16039 |
| chr11_-_110095937 | 2.10 |
ENSMUST00000106664.3
ENSMUST00000046223.7 ENSMUST00000106662.1 |
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr3_-_98509967 | 2.08 |
ENSMUST00000179429.1
|
Hsd3b4
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 4 |
| chr6_+_8259327 | 1.94 |
ENSMUST00000159378.1
|
Gm16039
|
predicted gene 16039 |
| chr17_-_45686899 | 1.93 |
ENSMUST00000156254.1
|
Tmem63b
|
transmembrane protein 63b |
| chr2_-_177925604 | 1.86 |
ENSMUST00000108934.2
ENSMUST00000081529.4 |
C330013J21Rik
|
RIKEN cDNA C330013J21 gene |
| chr2_+_23068168 | 1.85 |
ENSMUST00000028121.7
ENSMUST00000114523.2 ENSMUST00000144088.1 |
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr7_-_18432033 | 1.85 |
ENSMUST00000019291.6
|
Psg28
|
pregnancy-specific glycoprotein 28 |
| chr8_-_41054771 | 1.80 |
ENSMUST00000093534.4
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr18_+_3383223 | 1.76 |
ENSMUST00000162301.1
ENSMUST00000161317.1 |
Cul2
|
cullin 2 |
| chrX_-_44790179 | 1.72 |
ENSMUST00000060481.2
|
Dcaf12l1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr2_+_177464733 | 1.65 |
ENSMUST00000099002.3
ENSMUST00000108952.3 |
Gm14420
|
predicted gene 14420 |
| chr3_+_19957037 | 1.64 |
ENSMUST00000091309.5
ENSMUST00000108329.1 ENSMUST00000003714.6 |
Cp
|
ceruloplasmin |
| chr3_-_39359128 | 1.61 |
ENSMUST00000056409.2
|
Gm9845
|
predicted pseudogene 9845 |
| chr9_+_54538984 | 1.55 |
ENSMUST00000060242.5
ENSMUST00000118413.1 |
Sh2d7
|
SH2 domain containing 7 |
| chr17_+_84957993 | 1.53 |
ENSMUST00000080217.6
ENSMUST00000112304.2 |
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chr12_+_76255209 | 1.51 |
ENSMUST00000021443.5
|
Mthfd1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent), methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthase |
| chr13_-_86046901 | 1.51 |
ENSMUST00000131011.1
|
Cox7c
|
cytochrome c oxidase subunit VIIc |
| chr2_-_175175871 | 1.45 |
ENSMUST00000109054.2
|
Gm14443
|
predicted gene 14443 |
| chr13_+_60601921 | 1.44 |
ENSMUST00000077453.5
|
Dapk1
|
death associated protein kinase 1 |
| chr19_+_8920358 | 1.44 |
ENSMUST00000096243.5
|
B3gat3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr5_+_104170676 | 1.37 |
ENSMUST00000112771.1
|
Dspp
|
dentin sialophosphoprotein |
| chr3_+_19957088 | 1.35 |
ENSMUST00000108328.1
|
Cp
|
ceruloplasmin |
| chr11_-_110251736 | 1.34 |
ENSMUST00000044003.7
|
Abca6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr2_+_175275125 | 1.32 |
ENSMUST00000109051.1
|
Gm14440
|
predicted gene 14440 |
| chr5_+_144255223 | 1.32 |
ENSMUST00000056578.6
|
Bri3
|
brain protein I3 |
| chr3_-_28765364 | 1.30 |
ENSMUST00000094335.3
|
Gm6505
|
predicted pseudogene 6505 |
| chr19_-_3575708 | 1.30 |
ENSMUST00000113997.2
ENSMUST00000025846.8 ENSMUST00000172362.1 |
Ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr2_-_128967725 | 1.28 |
ENSMUST00000099385.2
|
Gm10762
|
predicted gene 10762 |
| chr6_-_143947061 | 1.26 |
ENSMUST00000124233.1
|
Sox5
|
SRY-box containing gene 5 |
| chr7_+_119690026 | 1.26 |
ENSMUST00000047045.8
|
Acsm4
|
acyl-CoA synthetase medium-chain family member 4 |
| chr1_-_138842429 | 1.24 |
ENSMUST00000112026.2
ENSMUST00000019374.7 |
Lhx9
|
LIM homeobox protein 9 |
| chr5_-_36988922 | 1.24 |
ENSMUST00000166339.1
ENSMUST00000043964.6 |
Wfs1
|
Wolfram syndrome 1 homolog (human) |
| chr5_-_113280572 | 1.23 |
ENSMUST00000112324.1
ENSMUST00000057209.5 |
Sgsm1
|
small G protein signaling modulator 1 |
| chr16_-_23029062 | 1.23 |
ENSMUST00000115349.2
|
Kng2
|
kininogen 2 |
| chrX_+_38600626 | 1.22 |
ENSMUST00000000365.2
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr4_+_145670685 | 1.20 |
ENSMUST00000105738.2
|
Gm13242
|
predicted gene 13242 |
| chr11_-_96829904 | 1.20 |
ENSMUST00000107657.1
|
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr16_-_23029012 | 1.19 |
ENSMUST00000039338.6
|
Kng2
|
kininogen 2 |
| chr11_-_31671727 | 1.19 |
ENSMUST00000109415.1
|
Bod1
|
biorientation of chromosomes in cell division 1 |
| chr10_-_49788743 | 1.19 |
ENSMUST00000105483.1
ENSMUST00000105487.1 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr16_-_37384915 | 1.18 |
ENSMUST00000114787.1
ENSMUST00000114782.1 ENSMUST00000023526.2 ENSMUST00000114775.1 |
Stxbp5l
|
syntaxin binding protein 5-like |
| chr9_-_108263706 | 1.18 |
ENSMUST00000171412.1
|
Dag1
|
dystroglycan 1 |
| chr16_-_23029080 | 1.17 |
ENSMUST00000100046.2
|
Kng2
|
kininogen 2 |
| chr4_+_147492417 | 1.15 |
ENSMUST00000105721.2
|
Gm13152
|
predicted gene 13152 |
| chr11_-_96829959 | 1.15 |
ENSMUST00000081775.5
|
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr9_-_111271568 | 1.13 |
ENSMUST00000035079.3
|
Mlh1
|
mutL homolog 1 (E. coli) |
| chr9_+_119341294 | 1.13 |
ENSMUST00000039784.5
|
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr11_+_5520652 | 1.12 |
ENSMUST00000063084.9
|
Xbp1
|
X-box binding protein 1 |
| chr11_+_58421103 | 1.12 |
ENSMUST00000013797.2
|
1810065E05Rik
|
RIKEN cDNA 1810065E05 gene |
| chrX_-_36874111 | 1.11 |
ENSMUST00000047486.5
|
C330007P06Rik
|
RIKEN cDNA C330007P06 gene |
| chr2_-_101621033 | 1.09 |
ENSMUST00000090513.4
|
B230118H07Rik
|
RIKEN cDNA B230118H07 gene |
| chr16_-_37384940 | 1.07 |
ENSMUST00000114781.1
ENSMUST00000114780.1 |
Stxbp5l
|
syntaxin binding protein 5-like |
| chr7_-_144939823 | 1.06 |
ENSMUST00000093962.4
|
Ccnd1
|
cyclin D1 |
| chr9_-_104102550 | 1.05 |
ENSMUST00000050139.4
|
Ackr4
|
atypical chemokine receptor 4 |
| chr7_-_26394238 | 1.02 |
ENSMUST00000041845.7
ENSMUST00000085944.5 |
Nlrp9c
|
NLR family, pyrin domain containing 9C |
| chr3_-_65392579 | 1.02 |
ENSMUST00000029414.5
|
Ssr3
|
signal sequence receptor, gamma |
| chr10_-_77799133 | 1.02 |
ENSMUST00000178581.1
|
Gm19668
|
predicted gene, 19668 |
| chr1_-_80213931 | 1.00 |
ENSMUST00000058748.1
|
Fam124b
|
family with sequence similarity 124, member B |
| chr1_+_60908993 | 1.00 |
ENSMUST00000027164.2
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr3_-_151749877 | 0.98 |
ENSMUST00000029671.7
|
Ifi44
|
interferon-induced protein 44 |
| chr18_-_73754457 | 0.93 |
ENSMUST00000041138.2
|
Elac1
|
elaC homolog 1 (E. coli) |
| chr2_-_92540967 | 0.92 |
ENSMUST00000142249.2
|
Gm13817
|
predicted gene 13817 |
| chr8_-_84237042 | 0.89 |
ENSMUST00000039480.5
|
Zswim4
|
zinc finger SWIM-type containing 4 |
| chr11_+_70525361 | 0.88 |
ENSMUST00000018430.6
|
Psmb6
|
proteasome (prosome, macropain) subunit, beta type 6 |
| chr8_+_90828820 | 0.88 |
ENSMUST00000109614.2
ENSMUST00000048665.6 |
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr19_-_41263931 | 0.88 |
ENSMUST00000025989.8
|
Tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr2_-_121442574 | 0.88 |
ENSMUST00000116432.1
|
Ell3
|
elongation factor RNA polymerase II-like 3 |
| chr13_+_60602182 | 0.88 |
ENSMUST00000044083.7
|
Dapk1
|
death associated protein kinase 1 |
| chrX_-_122397351 | 0.88 |
ENSMUST00000079490.4
|
Nap1l3
|
nucleosome assembly protein 1-like 3 |
| chr19_-_32061438 | 0.85 |
ENSMUST00000096119.4
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr11_-_110095974 | 0.84 |
ENSMUST00000100287.2
|
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr4_-_150003130 | 0.84 |
ENSMUST00000084117.6
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chrX_+_37831686 | 0.84 |
ENSMUST00000096454.1
|
Rhox7
|
reproductive homeobox 7 |
| chr6_-_143947092 | 0.84 |
ENSMUST00000144289.1
ENSMUST00000111748.1 |
Sox5
|
SRY-box containing gene 5 |
| chr2_-_132578244 | 0.84 |
ENSMUST00000110142.1
|
Gpcpd1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
| chr11_+_108381164 | 0.84 |
ENSMUST00000146050.1
ENSMUST00000152958.1 |
Apoh
|
apolipoprotein H |
| chr12_-_83921899 | 0.77 |
ENSMUST00000117217.1
|
Numb
|
numb gene homolog (Drosophila) |
| chr15_-_5108492 | 0.77 |
ENSMUST00000118365.2
|
Card6
|
caspase recruitment domain family, member 6 |
| chr2_+_59484645 | 0.76 |
ENSMUST00000028369.5
|
Dapl1
|
death associated protein-like 1 |
| chr17_+_21690766 | 0.76 |
ENSMUST00000097384.1
|
Gm10509
|
predicted gene 10509 |
| chr9_+_75051977 | 0.74 |
ENSMUST00000170310.1
ENSMUST00000166549.1 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr9_+_44398176 | 0.72 |
ENSMUST00000165839.1
|
Slc37a4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr15_-_73819410 | 0.72 |
ENSMUST00000110021.2
|
Mroh5
|
maestro heat-like repeat family member 5 |
| chr9_+_7571396 | 0.71 |
ENSMUST00000120900.1
ENSMUST00000093896.3 ENSMUST00000151853.1 ENSMUST00000152878.1 |
Mmp27
|
matrix metallopeptidase 27 |
| chrX_-_70365052 | 0.70 |
ENSMUST00000101509.2
|
Ids
|
iduronate 2-sulfatase |
| chr2_+_32535315 | 0.68 |
ENSMUST00000133512.1
ENSMUST00000048375.5 |
Fam102a
|
family with sequence similarity 102, member A |
| chr4_+_98546710 | 0.68 |
ENSMUST00000102792.3
|
Inadl
|
InaD-like (Drosophila) |
| chr3_-_152982240 | 0.67 |
ENSMUST00000044278.5
|
St6galnac5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr4_+_42949814 | 0.67 |
ENSMUST00000037872.3
ENSMUST00000098112.2 |
Dnajb5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr18_+_77938452 | 0.67 |
ENSMUST00000044622.5
|
Epg5
|
ectopic P-granules autophagy protein 5 homolog (C. elegans) |
| chr1_+_134560190 | 0.65 |
ENSMUST00000112198.1
ENSMUST00000112197.1 |
Kdm5b
|
lysine (K)-specific demethylase 5B |
| chrX_-_37897232 | 0.65 |
ENSMUST00000115156.1
|
Gm14543
|
predicted gene 14543 |
| chr2_+_55411790 | 0.65 |
ENSMUST00000155997.1
ENSMUST00000128307.1 |
Gm14033
|
predicted gene 14033 |
| chr17_-_24886304 | 0.64 |
ENSMUST00000044252.5
|
Nubp2
|
nucleotide binding protein 2 |
| chr16_-_93603803 | 0.62 |
ENSMUST00000023669.5
ENSMUST00000113951.2 |
Setd4
|
SET domain containing 4 |
| chr11_-_79296906 | 0.62 |
ENSMUST00000068448.2
|
Gm9964
|
predicted gene 9964 |
| chr12_-_87644467 | 0.62 |
ENSMUST00000180181.1
|
Gm16381
|
predicted gene 16381 |
| chr1_+_134560157 | 0.61 |
ENSMUST00000047714.7
|
Kdm5b
|
lysine (K)-specific demethylase 5B |
| chr17_+_75435886 | 0.60 |
ENSMUST00000164192.1
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr15_-_101562889 | 0.60 |
ENSMUST00000023714.4
|
4732456N10Rik
|
RIKEN cDNA 4732456N10 gene |
| chr14_+_63046988 | 0.60 |
ENSMUST00000067990.1
ENSMUST00000111203.1 |
Defb42
|
defensin beta 42 |
| chr13_-_43171461 | 0.59 |
ENSMUST00000179852.1
ENSMUST00000021797.8 |
Tbc1d7
|
TBC1 domain family, member 7 |
| chr1_+_164275559 | 0.58 |
ENSMUST00000027867.6
|
Ccdc181
|
coiled-coil domain containing 181 |
| chr4_-_43653542 | 0.58 |
ENSMUST00000084646.4
|
Spag8
|
sperm associated antigen 8 |
| chr3_+_20057802 | 0.57 |
ENSMUST00000002502.5
|
Hltf
|
helicase-like transcription factor |
| chr10_+_78425670 | 0.57 |
ENSMUST00000005185.6
|
Cstb
|
cystatin B |
| chr7_+_99735145 | 0.57 |
ENSMUST00000098264.1
|
Olfr520
|
olfactory receptor 520 |
| chr9_-_100546053 | 0.56 |
ENSMUST00000116522.1
|
Nck1
|
non-catalytic region of tyrosine kinase adaptor protein 1 |
| chr14_+_30825580 | 0.56 |
ENSMUST00000006701.5
|
Tmem110
|
transmembrane protein 110 |
| chr17_-_53539411 | 0.55 |
ENSMUST00000056198.3
|
Pp2d1
|
protein phosphatase 2C-like domain containing 1 |
| chr4_+_98546919 | 0.54 |
ENSMUST00000030290.7
|
Inadl
|
InaD-like (Drosophila) |
| chr7_-_28050028 | 0.54 |
ENSMUST00000032824.9
|
Psmc4
|
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| chr13_+_3538075 | 0.54 |
ENSMUST00000059515.6
|
Gdi2
|
guanosine diphosphate (GDP) dissociation inhibitor 2 |
| chr7_+_15882617 | 0.51 |
ENSMUST00000125993.1
ENSMUST00000130566.1 |
Crxos1
|
Crx opposite strand transcript 1 |
| chr4_-_59138983 | 0.50 |
ENSMUST00000107547.1
|
AI481877
|
expressed sequence AI481877 |
| chrX_-_134541847 | 0.50 |
ENSMUST00000054213.4
|
Timm8a1
|
translocase of inner mitochondrial membrane 8A1 |
| chr3_+_69131233 | 0.50 |
ENSMUST00000177708.1
|
Gm1647
|
predicted gene 1647 |
| chr11_-_72215592 | 0.50 |
ENSMUST00000021157.8
|
Med31
|
mediator of RNA polymerase II transcription, subunit 31 homolog (yeast) |
| chr12_-_83921809 | 0.48 |
ENSMUST00000135962.1
ENSMUST00000155112.1 ENSMUST00000136848.1 ENSMUST00000126943.1 |
Numb
|
numb gene homolog (Drosophila) |
| chr4_+_146156824 | 0.48 |
ENSMUST00000168483.2
|
Zfp600
|
zinc finger protein 600 |
| chr7_+_76411062 | 0.47 |
ENSMUST00000026854.3
ENSMUST00000107442.2 |
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr6_+_115853470 | 0.47 |
ENSMUST00000112925.1
ENSMUST00000112923.1 ENSMUST00000038234.6 |
Ift122
|
intraflagellar transport 122 |
| chr17_+_21691860 | 0.47 |
ENSMUST00000072133.4
|
Gm10226
|
predicted gene 10226 |
| chr1_-_56972437 | 0.47 |
ENSMUST00000042857.7
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr4_+_147021850 | 0.46 |
ENSMUST00000075775.5
|
Rex2
|
reduced expression 2 |
| chr4_-_118809814 | 0.46 |
ENSMUST00000105035.1
ENSMUST00000084313.3 |
Olfr1335
|
olfactory receptor 1335 |
| chr1_+_5588493 | 0.45 |
ENSMUST00000160777.1
ENSMUST00000027038.4 |
Oprk1
|
opioid receptor, kappa 1 |
| chr15_-_5108469 | 0.44 |
ENSMUST00000141020.1
|
Card6
|
caspase recruitment domain family, member 6 |
| chr3_-_37125943 | 0.44 |
ENSMUST00000029275.5
|
Il2
|
interleukin 2 |
| chr4_+_146514920 | 0.43 |
ENSMUST00000140089.1
ENSMUST00000179175.1 |
Gm13247
|
predicted gene 13247 |
| chr3_+_146117451 | 0.43 |
ENSMUST00000140214.1
|
Mcoln3
|
mucolipin 3 |
| chr3_-_135691512 | 0.42 |
ENSMUST00000029812.7
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
| chr19_+_11912389 | 0.42 |
ENSMUST00000061618.7
|
Patl1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr11_-_99422252 | 0.41 |
ENSMUST00000017741.3
|
Krt12
|
keratin 12 |
| chr4_+_146971976 | 0.40 |
ENSMUST00000146688.2
|
Gm13150
|
predicted gene 13150 |
| chr1_-_130629612 | 0.39 |
ENSMUST00000142416.1
ENSMUST00000039862.4 ENSMUST00000128128.1 |
Zp3r
|
zona pellucida 3 receptor |
| chr11_+_58665561 | 0.39 |
ENSMUST00000072030.3
|
Olfr322
|
olfactory receptor 322 |
| chr4_-_43653560 | 0.38 |
ENSMUST00000107870.2
|
Spag8
|
sperm associated antigen 8 |
| chr16_+_32332238 | 0.38 |
ENSMUST00000115151.3
|
Ubxn7
|
UBX domain protein 7 |
| chr2_+_128862947 | 0.38 |
ENSMUST00000110324.1
|
Fbln7
|
fibulin 7 |
| chr18_+_14424821 | 0.37 |
ENSMUST00000069552.5
|
Gm5160
|
predicted gene 5160 |
| chr15_-_79774408 | 0.37 |
ENSMUST00000023055.6
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr19_-_11856001 | 0.36 |
ENSMUST00000079875.3
|
Olfr1418
|
olfactory receptor 1418 |
| chr15_-_79774383 | 0.35 |
ENSMUST00000069877.5
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr9_-_80465429 | 0.35 |
ENSMUST00000085289.5
ENSMUST00000185068.1 ENSMUST00000113250.3 |
Impg1
|
interphotoreceptor matrix proteoglycan 1 |
| chr1_+_60909148 | 0.34 |
ENSMUST00000097720.3
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr8_-_54724317 | 0.34 |
ENSMUST00000129132.2
ENSMUST00000150488.1 ENSMUST00000127511.2 |
Wdr17
|
WD repeat domain 17 |
| chr18_-_53744509 | 0.33 |
ENSMUST00000049811.6
|
Cep120
|
centrosomal protein 120 |
| chrX_-_92599572 | 0.33 |
ENSMUST00000113955.1
|
Mageb18
|
melanoma antigen family B, 18 |
| chr1_+_169928648 | 0.32 |
ENSMUST00000094348.3
|
1700084C01Rik
|
RIKEN cDNA 1700084C01 gene |
| chr4_+_150997081 | 0.32 |
ENSMUST00000030803.1
|
Uts2
|
urotensin 2 |
| chr6_-_23655129 | 0.30 |
ENSMUST00000104979.1
|
Rnf148
|
ring finger protein 148 |
| chrX_+_73064787 | 0.30 |
ENSMUST00000060418.6
|
Pnma3
|
paraneoplastic antigen MA3 |
| chr13_+_22293193 | 0.30 |
ENSMUST00000091735.1
|
Vmn1r196
|
vomeronasal 1 receptor 196 |
| chrM_+_11734 | 0.28 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr11_-_115708538 | 0.28 |
ENSMUST00000106495.1
ENSMUST00000021090.7 |
Grb2
|
growth factor receptor bound protein 2 |
| chr14_+_55015454 | 0.28 |
ENSMUST00000022815.8
|
Ngdn
|
neuroguidin, EIF4E binding protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pbx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.9 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 2.2 | 13.2 | GO:0008355 | olfactory learning(GO:0008355) |
| 1.6 | 4.9 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 1.2 | 3.6 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.9 | 2.6 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.6 | 1.3 | GO:2000864 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.6 | 2.4 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.6 | 2.3 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.5 | 3.2 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.5 | 3.8 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.5 | 2.3 | GO:0045994 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.5 | 1.4 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.4 | 1.3 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.4 | 1.2 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.4 | 5.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.4 | 10.9 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.4 | 1.1 | GO:0071332 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) cellular response to fructose stimulus(GO:0071332) |
| 0.3 | 1.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 1.2 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.3 | 3.5 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 1.4 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.3 | 1.1 | GO:0051311 | male meiosis chromosome segregation(GO:0007060) meiotic metaphase plate congression(GO:0051311) |
| 0.3 | 1.8 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 1.5 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.3 | 1.5 | GO:0009256 | homoserine metabolic process(GO:0009092) 10-formyltetrahydrofolate metabolic process(GO:0009256) transsulfuration(GO:0019346) tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.2 | 1.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.2 | 2.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.2 | 1.3 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.2 | 0.8 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 | 0.6 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.2 | 0.9 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.2 | 2.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.2 | 9.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.2 | 2.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.2 | 1.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 | 0.9 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.7 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) post-embryonic hemopoiesis(GO:0035166) |
| 0.1 | 0.5 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.4 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) |
| 0.1 | 0.4 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 1.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.4 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.1 | 0.3 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 3.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 11.9 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 3.0 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 6.5 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 1.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.5 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 0.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 3.6 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 1.0 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.7 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.8 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 1.8 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 0.5 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 2.2 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.1 | 0.6 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 0.2 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.0 | 0.8 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 2.3 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 2.3 | GO:0001662 | behavioral fear response(GO:0001662) |
| 0.0 | 0.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.4 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 1.1 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.6 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.3 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 3.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 6.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 1.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.7 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 4.1 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.6 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 3.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 4.4 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 1.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.6 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 3.3 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.8 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.5 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 1.3 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 1.2 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 0.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.2 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 1.8 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 22.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.4 | 3.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.4 | 1.1 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.4 | 1.8 | GO:0030891 | VCB complex(GO:0030891) |
| 0.3 | 2.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 2.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 1.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 3.8 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 2.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 3.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.4 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 1.2 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.1 | 0.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.5 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 0.6 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.5 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 3.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 1.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 2.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 1.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 13.8 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 3.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 3.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 4.1 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 1.3 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 24.9 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 3.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.7 | GO:0030286 | dynein complex(GO:0030286) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.2 | GO:0005186 | pheromone activity(GO:0005186) |
| 3.6 | 10.9 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 1.0 | 2.9 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.9 | 11.9 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.8 | 6.5 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.5 | 18.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.5 | 1.5 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.4 | 2.6 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.4 | 2.4 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.4 | 1.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.3 | 12.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 1.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.3 | 3.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 2.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.3 | 3.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 3.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 14.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.3 | 0.8 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.2 | 8.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 2.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 0.8 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.2 | 1.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 1.1 | GO:0008410 | acetyl-CoA C-acetyltransferase activity(GO:0003985) CoA-transferase activity(GO:0008410) palmitoyl-CoA oxidase activity(GO:0016401) C-acetyltransferase activity(GO:0016453) |
| 0.2 | 0.7 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 1.3 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.2 | 5.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 5.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.9 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 2.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.5 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 3.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.6 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 1.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 2.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 4.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 1.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.8 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 4.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 2.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.7 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 1.0 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 2.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.9 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 4.2 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 2.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.1 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 3.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 2.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 2.4 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.2 | GO:0043855 | cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 3.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.6 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 1.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 1.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 2.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 5.9 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 2.3 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 3.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 3.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 2.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | SIG CHEMOTAXIS | Genes related to chemotaxis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 10.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.6 | 9.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.4 | 5.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 6.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 2.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 3.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 3.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 3.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 2.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.1 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.1 | 1.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.3 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 1.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 3.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.4 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 2.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.6 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |