Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
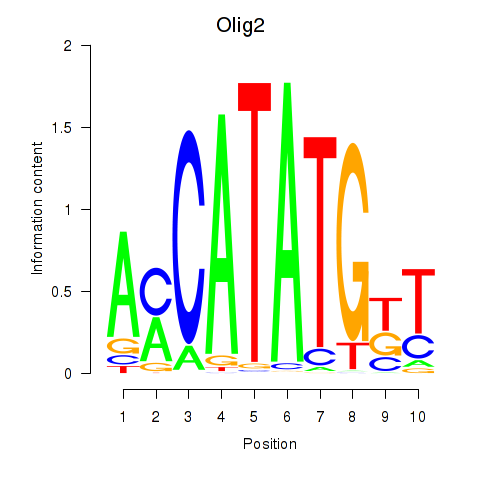
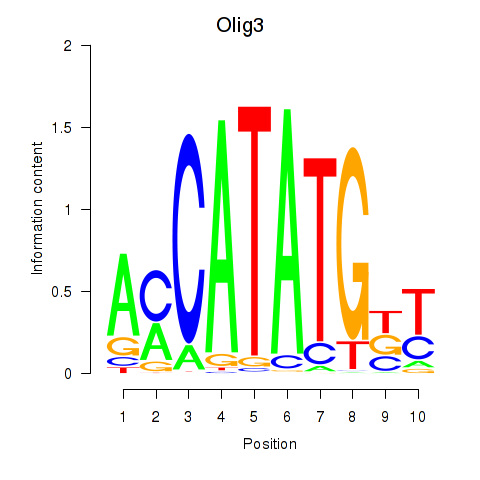
Results for Olig2_Olig3
Z-value: 1.46
Transcription factors associated with Olig2_Olig3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Olig2
|
ENSMUSG00000039830.8 | oligodendrocyte transcription factor 2 |
|
Olig3
|
ENSMUSG00000045591.5 | oligodendrocyte transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Olig2 | mm10_v2_chr16_+_91225550_91225579 | -0.37 | 2.5e-02 | Click! |
| Olig3 | mm10_v2_chr10_+_19356558_19356565 | -0.29 | 9.0e-02 | Click! |
Activity profile of Olig2_Olig3 motif
Sorted Z-values of Olig2_Olig3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_20601958 | 16.45 |
ENSMUST00000087638.3
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr11_+_48838672 | 13.92 |
ENSMUST00000129674.1
|
Trim7
|
tripartite motif-containing 7 |
| chr10_+_87861309 | 11.16 |
ENSMUST00000122100.1
|
Igf1
|
insulin-like growth factor 1 |
| chr6_+_121838514 | 10.81 |
ENSMUST00000032228.8
|
Mug1
|
murinoglobulin 1 |
| chr6_+_122006798 | 10.78 |
ENSMUST00000081777.6
|
Mug2
|
murinoglobulin 2 |
| chr5_-_87337165 | 10.46 |
ENSMUST00000031195.2
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr5_-_87092546 | 8.63 |
ENSMUST00000132667.1
ENSMUST00000145617.1 ENSMUST00000094649.4 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr15_-_76126538 | 8.48 |
ENSMUST00000054022.5
ENSMUST00000089654.3 |
BC024139
|
cDNA sequence BC024139 |
| chr4_-_61782222 | 8.14 |
ENSMUST00000107477.1
ENSMUST00000080606.2 |
Mup19
|
major urinary protein 19 |
| chr9_-_48605147 | 7.51 |
ENSMUST00000034808.5
ENSMUST00000119426.1 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr8_+_85492568 | 7.44 |
ENSMUST00000034136.5
|
Gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr8_-_5105232 | 7.07 |
ENSMUST00000023835.1
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr2_-_52558539 | 6.74 |
ENSMUST00000102760.3
ENSMUST00000102761.2 |
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr8_+_105048592 | 6.74 |
ENSMUST00000093222.5
ENSMUST00000093223.3 |
Ces3a
|
carboxylesterase 3A |
| chr7_-_139582790 | 6.24 |
ENSMUST00000106095.2
|
Nkx6-2
|
NK6 homeobox 2 |
| chr2_-_25500613 | 5.78 |
ENSMUST00000040042.4
|
C8g
|
complement component 8, gamma polypeptide |
| chr5_-_147322435 | 5.75 |
ENSMUST00000100433.4
|
Urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5) decarboxylase |
| chr1_-_166309585 | 5.74 |
ENSMUST00000168347.1
|
5330438I03Rik
|
RIKEN cDNA 5330438I03 gene |
| chr5_-_87254804 | 5.62 |
ENSMUST00000075858.3
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr17_-_32420965 | 5.29 |
ENSMUST00000170392.1
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr5_-_87091150 | 4.98 |
ENSMUST00000154455.1
|
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr11_-_69805617 | 4.96 |
ENSMUST00000051025.4
|
Tmem102
|
transmembrane protein 102 |
| chr11_-_73324616 | 4.87 |
ENSMUST00000021119.2
|
Aspa
|
aspartoacylase |
| chr3_-_107943705 | 4.64 |
ENSMUST00000106680.1
ENSMUST00000106684.1 ENSMUST00000106685.2 |
Gstm6
|
glutathione S-transferase, mu 6 |
| chr16_-_46010212 | 4.50 |
ENSMUST00000130481.1
|
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr19_+_26753588 | 4.45 |
ENSMUST00000177116.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_-_180199587 | 4.40 |
ENSMUST00000161743.1
|
Adck3
|
aarF domain containing kinase 3 |
| chr9_-_121916288 | 4.36 |
ENSMUST00000062474.4
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr18_+_12643329 | 4.34 |
ENSMUST00000025294.7
|
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr6_-_87690819 | 4.31 |
ENSMUST00000162547.1
|
1810020O05Rik
|
Riken cDNA 1810020O05 gene |
| chr1_-_139560158 | 4.29 |
ENSMUST00000160423.1
ENSMUST00000023965.5 |
Cfhr1
|
complement factor H-related 1 |
| chr4_+_86053887 | 4.19 |
ENSMUST00000107178.2
ENSMUST00000048885.5 ENSMUST00000141889.1 ENSMUST00000120678.1 |
Adamtsl1
|
ADAMTS-like 1 |
| chrM_+_5319 | 4.06 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I |
| chr3_+_19985612 | 4.05 |
ENSMUST00000172860.1
|
Cp
|
ceruloplasmin |
| chr1_+_167618246 | 3.89 |
ENSMUST00000111380.1
|
Rxrg
|
retinoid X receptor gamma |
| chr19_-_20727533 | 3.84 |
ENSMUST00000025656.3
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr1_-_186117251 | 3.84 |
ENSMUST00000045388.7
|
Lyplal1
|
lysophospholipase-like 1 |
| chr8_+_94838321 | 3.72 |
ENSMUST00000034234.8
ENSMUST00000159871.1 |
Coq9
|
coenzyme Q9 homolog (yeast) |
| chr16_+_43235856 | 3.66 |
ENSMUST00000146708.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr2_+_155382186 | 3.57 |
ENSMUST00000134218.1
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chrX_+_141475385 | 3.56 |
ENSMUST00000112931.1
ENSMUST00000112930.1 |
Col4a5
|
collagen, type IV, alpha 5 |
| chr9_+_72985568 | 3.41 |
ENSMUST00000150826.2
ENSMUST00000085350.4 ENSMUST00000140675.1 |
Ccpg1
|
cell cycle progression 1 |
| chr17_+_24878724 | 3.37 |
ENSMUST00000050714.6
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr1_-_139781236 | 3.37 |
ENSMUST00000027612.8
ENSMUST00000111989.2 ENSMUST00000111986.2 |
Gm4788
|
predicted gene 4788 |
| chr14_+_32991392 | 3.35 |
ENSMUST00000120866.1
ENSMUST00000120588.1 |
Lrrc18
|
leucine rich repeat containing 18 |
| chr1_-_139858684 | 3.29 |
ENSMUST00000094489.3
|
Cfhr2
|
complement factor H-related 2 |
| chr9_-_78322357 | 3.29 |
ENSMUST00000095071.4
|
Gm8074
|
predicted gene 8074 |
| chr5_-_24447587 | 3.24 |
ENSMUST00000127194.1
ENSMUST00000115033.1 ENSMUST00000123167.1 ENSMUST00000030799.8 |
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr16_+_44867097 | 3.21 |
ENSMUST00000102805.3
|
Cd200r2
|
Cd200 receptor 2 |
| chr8_+_22283441 | 3.16 |
ENSMUST00000077194.1
|
Tpte
|
transmembrane phosphatase with tensin homology |
| chr16_+_11008898 | 3.09 |
ENSMUST00000180624.1
|
Gm4262
|
predicted gene 4262 |
| chr6_+_90462562 | 3.09 |
ENSMUST00000032174.5
|
Klf15
|
Kruppel-like factor 15 |
| chr2_+_69670100 | 3.04 |
ENSMUST00000100050.3
|
Klhl41
|
kelch-like 41 |
| chr11_-_69920581 | 3.01 |
ENSMUST00000108610.1
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr14_+_32991430 | 3.01 |
ENSMUST00000123822.1
ENSMUST00000120951.1 |
Lrrc18
|
leucine rich repeat containing 18 |
| chr8_+_36489191 | 2.96 |
ENSMUST00000171777.1
|
6430573F11Rik
|
RIKEN cDNA 6430573F11 gene |
| chr2_+_137663424 | 2.95 |
ENSMUST00000134833.1
|
Gm14064
|
predicted gene 14064 |
| chr9_+_72985504 | 2.89 |
ENSMUST00000156879.1
|
Ccpg1
|
cell cycle progression 1 |
| chr6_-_129233969 | 2.86 |
ENSMUST00000181517.1
|
2310001H17Rik
|
RIKEN cDNA 2310001H17 gene |
| chr9_+_66946057 | 2.78 |
ENSMUST00000040917.7
ENSMUST00000127896.1 |
Rps27l
|
ribosomal protein S27-like |
| chr10_+_110920170 | 2.75 |
ENSMUST00000020403.5
|
Csrp2
|
cysteine and glycine-rich protein 2 |
| chr2_-_86347764 | 2.71 |
ENSMUST00000099894.2
|
Olfr1055
|
olfactory receptor 1055 |
| chr5_-_103977326 | 2.64 |
ENSMUST00000120320.1
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr5_-_45450221 | 2.58 |
ENSMUST00000015950.5
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr1_+_88166004 | 2.58 |
ENSMUST00000097659.4
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr16_-_46155077 | 2.50 |
ENSMUST00000059524.5
|
Gm4737
|
predicted gene 4737 |
| chr5_+_144255223 | 2.49 |
ENSMUST00000056578.6
|
Bri3
|
brain protein I3 |
| chr1_+_160978576 | 2.47 |
ENSMUST00000064725.5
|
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr10_-_31445921 | 2.45 |
ENSMUST00000000305.5
|
Tpd52l1
|
tumor protein D52-like 1 |
| chr1_-_74749221 | 2.45 |
ENSMUST00000081636.6
|
Prkag3
|
protein kinase, AMP-activated, gamma 3 non-catatlytic subunit |
| chr14_+_74732297 | 2.44 |
ENSMUST00000022573.10
ENSMUST00000175712.1 |
Esd
|
esterase D/formylglutathione hydrolase |
| chr3_-_123236134 | 2.42 |
ENSMUST00000106427.1
ENSMUST00000106426.1 ENSMUST00000051443.5 |
Synpo2
|
synaptopodin 2 |
| chr14_+_74732384 | 2.41 |
ENSMUST00000176957.1
|
Esd
|
esterase D/formylglutathione hydrolase |
| chr10_+_3540240 | 2.33 |
ENSMUST00000019896.4
|
Iyd
|
iodotyrosine deiodinase |
| chr5_-_103977360 | 2.33 |
ENSMUST00000048118.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr5_-_103977404 | 2.31 |
ENSMUST00000112803.2
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr3_-_113574242 | 2.31 |
ENSMUST00000142505.2
|
Amy1
|
amylase 1, salivary |
| chr8_-_34146974 | 2.29 |
ENSMUST00000033910.8
|
Leprotl1
|
leptin receptor overlapping transcript-like 1 |
| chr7_+_30458280 | 2.29 |
ENSMUST00000126297.1
|
Nphs1
|
nephrosis 1, nephrin |
| chr14_+_32991379 | 2.26 |
ENSMUST00000038956.4
|
Lrrc18
|
leucine rich repeat containing 18 |
| chr7_+_107567445 | 2.24 |
ENSMUST00000120990.1
|
Olfml1
|
olfactomedin-like 1 |
| chr11_+_78194696 | 2.19 |
ENSMUST00000060539.6
|
Proca1
|
protein interacting with cyclin A1 |
| chr14_+_103070216 | 2.19 |
ENSMUST00000022721.6
|
Cln5
|
ceroid-lipofuscinosis, neuronal 5 |
| chr10_-_117746356 | 2.14 |
ENSMUST00000079041.5
|
Slc35e3
|
solute carrier family 35, member E3 |
| chr6_-_129237948 | 2.12 |
ENSMUST00000181238.1
ENSMUST00000180379.1 |
2310001H17Rik
|
RIKEN cDNA 2310001H17 gene |
| chr5_-_135962275 | 2.12 |
ENSMUST00000054895.3
|
Srcrb4d
|
scavenger receptor cysteine rich domain containing, group B (4 domains) |
| chr14_+_41151442 | 2.11 |
ENSMUST00000047095.2
|
Mbl1
|
mannose-binding lectin (protein A) 1 |
| chr8_-_25038875 | 2.10 |
ENSMUST00000084031.4
|
Htra4
|
HtrA serine peptidase 4 |
| chr10_-_127522428 | 2.08 |
ENSMUST00000026470.4
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chrX_+_10252305 | 2.08 |
ENSMUST00000049910.6
|
Otc
|
ornithine transcarbamylase |
| chr11_+_69991633 | 2.06 |
ENSMUST00000108592.1
|
Gabarap
|
gamma-aminobutyric acid receptor associated protein |
| chr3_-_107943362 | 2.05 |
ENSMUST00000106683.1
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr17_+_35126316 | 2.05 |
ENSMUST00000061859.6
|
D17H6S53E
|
DNA segment, Chr 17, human D6S53E |
| chr5_+_90460889 | 2.04 |
ENSMUST00000031314.8
|
Alb
|
albumin |
| chr7_+_100009914 | 2.03 |
ENSMUST00000107084.1
|
Chrdl2
|
chordin-like 2 |
| chr17_+_37193889 | 2.03 |
ENSMUST00000038844.6
|
Ubd
|
ubiquitin D |
| chr3_-_85746266 | 2.03 |
ENSMUST00000118408.1
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr7_+_100022099 | 2.02 |
ENSMUST00000144808.1
|
Chrdl2
|
chordin-like 2 |
| chr4_+_107067012 | 2.00 |
ENSMUST00000154283.1
|
Cyb5rl
|
cytochrome b5 reductase-like |
| chrX_+_38772671 | 1.99 |
ENSMUST00000050744.5
|
6030498E09Rik
|
RIKEN cDNA 6030498E09 gene |
| chr5_-_135962265 | 1.95 |
ENSMUST00000111150.1
|
Srcrb4d
|
scavenger receptor cysteine rich domain containing, group B (4 domains) |
| chr9_-_88719798 | 1.95 |
ENSMUST00000113110.3
|
Gm2382
|
predicted gene 2382 |
| chr11_+_78194734 | 1.95 |
ENSMUST00000108317.2
|
Proca1
|
protein interacting with cyclin A1 |
| chr7_+_123462274 | 1.94 |
ENSMUST00000033023.3
|
Aqp8
|
aquaporin 8 |
| chr17_-_25792284 | 1.91 |
ENSMUST00000072735.7
|
Fam173a
|
family with sequence similarity 173, member A |
| chr2_-_160327494 | 1.91 |
ENSMUST00000099127.2
|
Gm826
|
predicted gene 826 |
| chr15_+_31568851 | 1.91 |
ENSMUST00000070918.6
|
Cmbl
|
carboxymethylenebutenolidase-like (Pseudomonas) |
| chr6_+_113472276 | 1.89 |
ENSMUST00000147316.1
|
Il17rc
|
interleukin 17 receptor C |
| chr4_+_107066985 | 1.88 |
ENSMUST00000106758.1
ENSMUST00000145324.1 ENSMUST00000106760.1 |
Cyb5rl
|
cytochrome b5 reductase-like |
| chr14_+_51411001 | 1.88 |
ENSMUST00000022438.5
ENSMUST00000163019.1 ENSMUST00000159674.1 |
Vmn2r88
|
vomeronasal 2, receptor 88 |
| chr1_-_180245757 | 1.88 |
ENSMUST00000111104.1
|
Psen2
|
presenilin 2 |
| chr13_+_119623819 | 1.87 |
ENSMUST00000099241.2
|
Ccl28
|
chemokine (C-C motif) ligand 28 |
| chr2_-_136891363 | 1.87 |
ENSMUST00000028730.6
ENSMUST00000110089.2 |
Mkks
|
McKusick-Kaufman syndrome |
| chr8_-_84893887 | 1.84 |
ENSMUST00000003907.7
ENSMUST00000182458.1 ENSMUST00000109745.1 ENSMUST00000142748.1 |
Gcdh
|
glutaryl-Coenzyme A dehydrogenase |
| chr4_-_148160031 | 1.78 |
ENSMUST00000057907.3
|
Fbxo44
|
F-box protein 44 |
| chr3_-_37419566 | 1.76 |
ENSMUST00000138949.1
ENSMUST00000149449.1 ENSMUST00000108117.2 ENSMUST00000108118.2 ENSMUST00000099130.2 ENSMUST00000052645.6 |
Nudt6
|
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| chr7_-_101581161 | 1.75 |
ENSMUST00000063920.2
|
Art2b
|
ADP-ribosyltransferase 2b |
| chr2_+_127587214 | 1.75 |
ENSMUST00000028852.6
|
Mrps5
|
mitochondrial ribosomal protein S5 |
| chr2_-_101649501 | 1.69 |
ENSMUST00000078494.5
ENSMUST00000160722.1 ENSMUST00000160037.1 |
Rag1
B230118H07Rik
|
recombination activating gene 1 RIKEN cDNA B230118H07 gene |
| chr18_-_35627223 | 1.66 |
ENSMUST00000025212.5
|
Slc23a1
|
solute carrier family 23 (nucleobase transporters), member 1 |
| chr2_-_3366569 | 1.65 |
ENSMUST00000115087.2
ENSMUST00000027955.4 |
Olah
|
oleoyl-ACP hydrolase |
| chr17_+_34153072 | 1.64 |
ENSMUST00000114232.2
|
H2-DMb1
|
histocompatibility 2, class II, locus Mb1 |
| chr5_+_124483458 | 1.62 |
ENSMUST00000111453.1
|
Snrnp35
|
small nuclear ribonucleoprotein 35 (U11/U12) |
| chr5_-_86906937 | 1.62 |
ENSMUST00000031181.9
ENSMUST00000113333.1 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr18_-_7297901 | 1.57 |
ENSMUST00000081275.4
|
Armc4
|
armadillo repeat containing 4 |
| chr6_-_130386874 | 1.56 |
ENSMUST00000032288.4
|
Klra1
|
killer cell lectin-like receptor, subfamily A, member 1 |
| chr7_-_118584669 | 1.56 |
ENSMUST00000044195.4
|
Tmc7
|
transmembrane channel-like gene family 7 |
| chr3_-_106483435 | 1.56 |
ENSMUST00000164330.1
|
2010016I18Rik
|
RIKEN cDNA 2010016I18 gene |
| chr7_-_44816586 | 1.55 |
ENSMUST00000047356.8
|
Atf5
|
activating transcription factor 5 |
| chr9_+_37401897 | 1.55 |
ENSMUST00000115048.1
|
Robo4
|
roundabout homolog 4 (Drosophila) |
| chr3_-_154330543 | 1.53 |
ENSMUST00000184966.1
ENSMUST00000177846.2 |
Lhx8
|
LIM homeobox protein 8 |
| chr14_+_32028989 | 1.52 |
ENSMUST00000022460.4
|
Galnt15
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
| chrY_-_3410167 | 1.52 |
ENSMUST00000169382.2
|
Gm21704
|
predicted gene, 21704 |
| chr11_+_97029925 | 1.49 |
ENSMUST00000021249.4
|
Scrn2
|
secernin 2 |
| chr14_+_28511344 | 1.44 |
ENSMUST00000112272.1
|
Wnt5a
|
wingless-related MMTV integration site 5A |
| chr4_+_133518963 | 1.44 |
ENSMUST00000149807.1
ENSMUST00000042919.9 ENSMUST00000153811.1 ENSMUST00000105901.1 ENSMUST00000121797.1 |
1810019J16Rik
|
RIKEN cDNA 1810019J16 gene |
| chr11_-_3722189 | 1.43 |
ENSMUST00000102950.3
ENSMUST00000101632.3 |
Osbp2
|
oxysterol binding protein 2 |
| chr17_+_88626549 | 1.43 |
ENSMUST00000163588.1
ENSMUST00000064035.6 |
Ston1
|
stonin 1 |
| chr10_-_34096507 | 1.42 |
ENSMUST00000069125.6
|
Fam26e
|
family with sequence similarity 26, member E |
| chr9_-_99717259 | 1.41 |
ENSMUST00000112882.2
ENSMUST00000131922.1 |
Cldn18
|
claudin 18 |
| chr2_+_175283298 | 1.41 |
ENSMUST00000098998.3
|
Gm14440
|
predicted gene 14440 |
| chr7_+_127800844 | 1.40 |
ENSMUST00000106271.1
ENSMUST00000138432.1 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chrY_+_87118255 | 1.39 |
ENSMUST00000179970.1
|
Gm21477
|
predicted gene, 21477 |
| chrY_+_84098711 | 1.39 |
ENSMUST00000177775.1
|
Gm21095
|
predicted gene, 21095 |
| chr9_-_59353430 | 1.38 |
ENSMUST00000026265.6
|
Bbs4
|
Bardet-Biedl syndrome 4 (human) |
| chr1_-_181511451 | 1.38 |
ENSMUST00000058825.4
|
Ccdc121
|
coiled-coil domain containing 121 |
| chrY_+_54656891 | 1.38 |
ENSMUST00000179631.1
|
Gm21894
|
predicted gene, 21894 |
| chr11_+_117232254 | 1.38 |
ENSMUST00000106354.2
|
Sept9
|
septin 9 |
| chr4_-_41870612 | 1.37 |
ENSMUST00000179680.1
|
Gm21966
|
predicted gene, 21966 |
| chr5_-_66618636 | 1.37 |
ENSMUST00000162382.1
ENSMUST00000160870.1 ENSMUST00000087256.5 ENSMUST00000160103.1 ENSMUST00000162349.1 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr9_-_26999491 | 1.36 |
ENSMUST00000060513.7
ENSMUST00000120367.1 |
Acad8
|
acyl-Coenzyme A dehydrogenase family, member 8 |
| chrY_+_2830680 | 1.36 |
ENSMUST00000100360.3
ENSMUST00000171534.1 ENSMUST00000179404.1 |
Rbmy
Gm10256
|
RNA binding motif protein, Y chromosome predicted gene 10256 |
| chr9_+_107580117 | 1.35 |
ENSMUST00000093785.4
|
Nat6
|
N-acetyltransferase 6 |
| chr3_-_30269947 | 1.35 |
ENSMUST00000091270.2
|
Gm10258
|
predicted gene 10258 |
| chr11_-_26210553 | 1.34 |
ENSMUST00000101447.3
|
5730522E02Rik
|
RIKEN cDNA 5730522E02 gene |
| chr4_+_41762309 | 1.34 |
ENSMUST00000108042.2
|
Il11ra1
|
interleukin 11 receptor, alpha chain 1 |
| chr4_+_154856200 | 1.34 |
ENSMUST00000050220.2
|
Ttc34
|
tetratricopeptide repeat domain 34 |
| chrY_+_55215153 | 1.33 |
ENSMUST00000180249.1
|
Gm20931
|
predicted gene, 20931 |
| chrY_-_35130404 | 1.33 |
ENSMUST00000180170.1
|
Gm20855
|
predicted gene, 20855 |
| chr5_-_24902315 | 1.33 |
ENSMUST00000131486.1
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr9_-_78587968 | 1.32 |
ENSMUST00000117645.1
ENSMUST00000119213.1 ENSMUST00000052441.5 |
Slc17a5
|
solute carrier family 17 (anion/sugar transporter), member 5 |
| chr5_-_92042999 | 1.32 |
ENSMUST00000069937.4
ENSMUST00000086978.5 |
Cdkl2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase) |
| chr10_-_22149270 | 1.32 |
ENSMUST00000179054.1
ENSMUST00000069372.6 |
E030030I06Rik
|
RIKEN cDNA E030030I06 gene |
| chr17_+_46772635 | 1.32 |
ENSMUST00000071430.5
|
2310039H08Rik
|
RIKEN cDNA 2310039H08 gene |
| chr10_+_128267997 | 1.32 |
ENSMUST00000050901.2
|
Apof
|
apolipoprotein F |
| chr19_-_9559204 | 1.31 |
ENSMUST00000090527.3
|
Stxbp3b
|
syntaxin-binding protein 3B |
| chr3_+_122895072 | 1.31 |
ENSMUST00000023820.5
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr11_-_3774706 | 1.31 |
ENSMUST00000155197.1
|
Osbp2
|
oxysterol binding protein 2 |
| chr16_-_44333135 | 1.30 |
ENSMUST00000047446.6
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr1_-_150392719 | 1.30 |
ENSMUST00000006167.6
ENSMUST00000094477.2 ENSMUST00000097547.3 |
BC003331
|
cDNA sequence BC003331 |
| chr3_+_60031754 | 1.29 |
ENSMUST00000029325.3
|
Aadac
|
arylacetamide deacetylase (esterase) |
| chr7_-_37772868 | 1.29 |
ENSMUST00000176205.1
|
Zfp536
|
zinc finger protein 536 |
| chr4_-_108301045 | 1.29 |
ENSMUST00000043616.6
|
Zyg11b
|
zyg-ll family member B, cell cycle regulator |
| chr2_-_85678741 | 1.28 |
ENSMUST00000099919.2
|
Olfr1006
|
olfactory receptor 1006 |
| chrX_-_124135910 | 1.28 |
ENSMUST00000094491.3
|
Vmn2r121
|
vomeronasal 2, receptor 121 |
| chrY_+_79320997 | 1.26 |
ENSMUST00000178063.1
|
Gm20916
|
predicted gene, 20916 |
| chr7_-_48456331 | 1.26 |
ENSMUST00000094384.3
|
Mrgprb1
|
MAS-related GPR, member B1 |
| chr12_-_80968075 | 1.25 |
ENSMUST00000095572.4
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr16_+_16213318 | 1.24 |
ENSMUST00000162150.1
ENSMUST00000161342.1 ENSMUST00000039408.2 |
Pkp2
|
plakophilin 2 |
| chr8_+_53511695 | 1.24 |
ENSMUST00000033920.4
|
Aga
|
aspartylglucosaminidase |
| chr14_-_16249675 | 1.23 |
ENSMUST00000022311.4
|
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr3_-_79737794 | 1.21 |
ENSMUST00000078527.6
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr17_+_88626569 | 1.21 |
ENSMUST00000150023.1
|
Ston1
|
stonin 1 |
| chr16_-_44332925 | 1.19 |
ENSMUST00000136381.1
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chrX_+_7579666 | 1.19 |
ENSMUST00000115740.1
ENSMUST00000115739.1 |
Foxp3
|
forkhead box P3 |
| chr2_+_126034647 | 1.18 |
ENSMUST00000064794.7
|
Fgf7
|
fibroblast growth factor 7 |
| chr8_+_94525067 | 1.18 |
ENSMUST00000098489.4
|
Nlrc5
|
NLR family, CARD domain containing 5 |
| chr6_-_130231638 | 1.17 |
ENSMUST00000088011.4
ENSMUST00000112013.1 ENSMUST00000049304.7 |
Klra7
|
killer cell lectin-like receptor, subfamily A, member 7 |
| chr11_+_120672992 | 1.15 |
ENSMUST00000026135.8
|
Aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
| chr6_+_78380700 | 1.15 |
ENSMUST00000101272.1
|
Reg3a
|
regenerating islet-derived 3 alpha |
| chr1_-_74304386 | 1.13 |
ENSMUST00000016309.9
|
Tmbim1
|
transmembrane BAX inhibitor motif containing 1 |
| chr5_-_66618772 | 1.12 |
ENSMUST00000162994.1
ENSMUST00000159512.1 ENSMUST00000159786.1 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr11_+_94284652 | 1.11 |
ENSMUST00000178891.1
|
Gm21885
|
predicted gene, 21885 |
| chr4_-_155669171 | 1.10 |
ENSMUST00000103176.3
|
Mib2
|
mindbomb homolog 2 (Drosophila) |
| chr7_+_43712505 | 1.10 |
ENSMUST00000066834.6
|
Klk13
|
kallikrein related-peptidase 13 |
| chr12_+_87147703 | 1.10 |
ENSMUST00000063117.8
|
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
| chrY_+_51126655 | 1.08 |
ENSMUST00000180133.1
|
Gm21117
|
predicted gene, 21117 |
| chr6_-_29507946 | 1.08 |
ENSMUST00000101614.3
ENSMUST00000078112.6 |
Kcp
|
kielin/chordin-like protein |
| chr2_-_35061431 | 1.07 |
ENSMUST00000028233.3
|
Hc
|
hemolytic complement |
| chr2_+_160888156 | 1.07 |
ENSMUST00000109457.2
|
Lpin3
|
lipin 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Olig2_Olig3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 16.5 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.9 | 7.4 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 1.9 | 11.2 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 1.6 | 6.3 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 1.2 | 4.8 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 1.1 | 5.3 | GO:0032827 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.9 | 6.2 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.8 | 4.9 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.8 | 13.0 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.7 | 3.0 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.7 | 4.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.7 | 2.1 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.7 | 2.0 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.7 | 6.7 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.7 | 4.0 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.7 | 3.3 | GO:1902019 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.6 | 3.8 | GO:0042997 | protein depalmitoylation(GO:0002084) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) macromolecule depalmitoylation(GO:0098734) |
| 0.6 | 1.8 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.5 | 5.8 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.5 | 2.1 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.5 | 4.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.5 | 6.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.5 | 1.4 | GO:0061347 | cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.5 | 1.9 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.4 | 11.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.4 | 3.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.4 | 1.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.4 | 1.2 | GO:0060618 | nipple development(GO:0060618) |
| 0.4 | 1.2 | GO:0045077 | CD4-positive, CD25-positive, alpha-beta regulatory T cell lineage commitment(GO:0002362) peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) negative regulation of histone deacetylation(GO:0031064) negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.4 | 1.2 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.4 | 5.0 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.4 | 2.2 | GO:1904426 | positive regulation of GTP binding(GO:1904426) |
| 0.3 | 1.7 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) |
| 0.3 | 1.7 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.3 | 1.0 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.3 | 3.7 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.3 | 1.7 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.3 | 2.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.3 | 1.0 | GO:0034443 | regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.3 | 2.6 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.3 | 2.3 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.3 | 1.0 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.3 | 0.9 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.3 | 1.2 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.3 | 2.4 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.3 | 0.9 | GO:0071462 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) cellular response to water stimulus(GO:0071462) |
| 0.3 | 0.9 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.3 | 4.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 1.4 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.3 | 2.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.3 | 0.8 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.3 | 0.8 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.2 | 3.4 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.2 | 0.7 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.2 | 3.0 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.2 | 2.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 3.7 | GO:0006744 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) ubiquinone biosynthetic process(GO:0006744) |
| 0.2 | 1.4 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 0.6 | GO:0042697 | menopause(GO:0042697) |
| 0.2 | 0.8 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.2 | 0.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.2 | 2.5 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.2 | 3.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.2 | 2.1 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 1.9 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 0.7 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.2 | 2.8 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.2 | 0.5 | GO:1902071 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.2 | 0.9 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 1.2 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 4.1 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 1.2 | GO:0045345 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.1 | 0.9 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 2.0 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 1.1 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 0.6 | GO:2000292 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.1 | 3.1 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 3.5 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.6 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 0.1 | 8.9 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 1.8 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 0.9 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.1 | 5.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.6 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.5 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.1 | 0.6 | GO:0009446 | spermidine biosynthetic process(GO:0008295) putrescine biosynthetic process(GO:0009446) |
| 0.1 | 1.3 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.7 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.1 | 0.6 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.3 | GO:1904444 | transepithelial water transport(GO:0035377) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) regulation of establishment of Sertoli cell barrier(GO:1904444) positive regulation of establishment of Sertoli cell barrier(GO:1904446) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.1 | 2.0 | GO:2000778 | positive regulation of interleukin-6 secretion(GO:2000778) |
| 0.1 | 0.7 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 1.3 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.5 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 1.0 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 0.3 | GO:0000239 | pachytene(GO:0000239) |
| 0.1 | 4.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.4 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.1 | 0.9 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.8 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 1.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 0.7 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.5 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.4 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.1 | 1.5 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 2.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.1 | 0.8 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.5 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 1.5 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 0.5 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.5 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 2.3 | GO:0044062 | regulation of excretion(GO:0044062) |
| 0.1 | 0.2 | GO:1903679 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 | 1.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 4.7 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.1 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.6 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 1.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.1 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 0.2 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 0.8 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 0.7 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.1 | 0.3 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.4 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 | 1.4 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.8 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.3 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 3.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.3 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.3 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 4.1 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 11.8 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 1.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 2.4 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 1.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.9 | GO:0034204 | lipid translocation(GO:0034204) |
| 0.0 | 2.6 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.4 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.2 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.1 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 1.3 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 1.3 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.5 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.3 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.7 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 5.5 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.4 | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.0 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.3 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.2 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.2 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.6 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) |
| 0.0 | 0.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 1.3 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 0.6 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.1 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.0 | 1.6 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.4 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.1 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.5 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.3 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 2.0 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.0 | 0.6 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.2 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 3.2 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 1.0 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.3 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 1.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.7 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.6 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.4 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 1.1 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.3 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.5 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.6 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.2 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 1.2 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 0.2 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.3 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 14.5 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.6 | 1.9 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.6 | 6.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.6 | 3.6 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.5 | 2.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.5 | 1.9 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.4 | 4.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.3 | 1.7 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.2 | 4.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 2.2 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.2 | 1.5 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.2 | 4.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 10.5 | GO:0000800 | lateral element(GO:0000800) |
| 0.2 | 3.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 3.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 1.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 1.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 1.2 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 0.9 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 1.6 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 2.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 3.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.8 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 0.8 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 5.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.4 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 1.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 5.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.9 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.1 | 1.9 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.4 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 0.7 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 2.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.9 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.9 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.6 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 7.3 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 0.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.4 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.3 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 1.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 3.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 3.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.4 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.5 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 4.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.5 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 2.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.6 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 1.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 9.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 2.1 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 3.1 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 2.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 20.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 1.6 | 4.9 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 1.6 | 4.8 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 1.1 | 5.3 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 1.1 | 7.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.9 | 33.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.9 | 2.6 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.7 | 2.1 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.5 | 2.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.5 | 3.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.5 | 1.4 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.4 | 1.3 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.4 | 2.5 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.4 | 5.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.4 | 4.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 3.9 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.4 | 3.8 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.4 | 3.9 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.4 | 2.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.3 | 1.3 | GO:0019970 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.3 | 1.7 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.3 | 2.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.3 | 0.9 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.3 | 1.2 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.3 | 0.9 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) 5'-3' exoribonuclease activity(GO:0004534) |
| 0.3 | 11.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.3 | 7.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.3 | 0.8 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.3 | 0.8 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.3 | 4.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 1.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 1.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 1.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 0.9 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 2.8 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.2 | 3.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 1.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 1.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 4.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 5.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 24.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 1.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 0.7 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.2 | 0.8 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.2 | 0.5 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.2 | 7.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 0.6 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.2 | 0.6 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.7 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 4.1 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.9 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 2.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 2.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.9 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 1.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 1.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.6 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.7 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 2.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.9 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 2.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 5.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.8 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 2.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.3 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.4 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.7 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.1 | 0.5 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.3 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 1.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 1.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 1.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 4.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.5 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 1.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 2.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.9 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 1.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.9 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.4 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) |
| 0.1 | 1.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 6.4 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 1.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 0.7 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.1 | 4.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.6 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.2 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.1 | 0.6 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 1.7 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 3.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 2.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 3.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.7 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 3.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.0 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 7.8 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 1.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 1.6 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 2.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 4.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 7.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.0 | 0.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.9 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 1.9 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 2.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 3.0 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.6 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.6 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.3 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 8.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.1 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 3.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.4 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.6 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 8.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 3.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 2.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 2.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 4.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 3.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 16.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 1.2 | 10.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.5 | 5.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 3.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 5.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 2.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 2.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 5.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 3.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 1.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 4.0 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 1.9 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 8.2 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 1.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 4.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 1.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 0.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 3.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.0 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 2.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 0.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 7.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.2 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 1.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 2.8 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 2.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.6 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |