Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
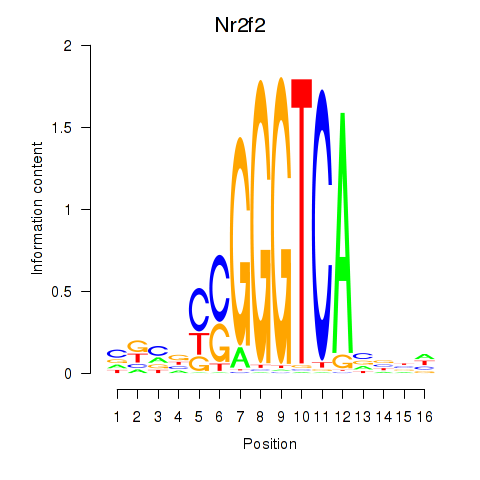
Results for Nr2f2
Z-value: 1.11
Transcription factors associated with Nr2f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2f2
|
ENSMUSG00000030551.7 | nuclear receptor subfamily 2, group F, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2f2 | mm10_v2_chr7_-_70360593_70360746 | 0.23 | 1.8e-01 | Click! |
Activity profile of Nr2f2 motif
Sorted Z-values of Nr2f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_105269837 | 9.00 |
ENSMUST00000172525.1
ENSMUST00000174837.1 ENSMUST00000173859.1 |
Hsf4
|
heat shock transcription factor 4 |
| chr8_+_105269788 | 8.70 |
ENSMUST00000036127.2
ENSMUST00000163734.2 |
Hsf4
|
heat shock transcription factor 4 |
| chr2_+_102658640 | 6.49 |
ENSMUST00000080210.3
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr7_-_30973464 | 5.59 |
ENSMUST00000001279.8
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr7_-_30973399 | 4.83 |
ENSMUST00000098553.4
ENSMUST00000147431.1 |
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr9_-_86695897 | 4.66 |
ENSMUST00000034989.8
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr7_-_30973367 | 4.60 |
ENSMUST00000108116.3
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chrX_-_75874536 | 4.47 |
ENSMUST00000033547.7
|
Pls3
|
plastin 3 (T-isoform) |
| chr7_-_99626936 | 4.10 |
ENSMUST00000178124.1
|
Gm4980
|
predicted gene 4980 |
| chr12_-_113260217 | 4.01 |
ENSMUST00000178282.1
|
Igha
|
immunoglobulin heavy constant alpha |
| chr11_+_72435511 | 3.94 |
ENSMUST00000076443.3
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr11_+_72435565 | 3.83 |
ENSMUST00000100903.2
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr10_-_128673896 | 3.74 |
ENSMUST00000054764.7
|
Suox
|
sulfite oxidase |
| chr17_-_56121946 | 3.74 |
ENSMUST00000041357.7
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr11_+_72435534 | 3.69 |
ENSMUST00000108499.1
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr3_+_121723515 | 3.33 |
ENSMUST00000029771.8
|
F3
|
coagulation factor III |
| chr11_-_5915124 | 3.30 |
ENSMUST00000109823.2
ENSMUST00000109822.1 |
Gck
|
glucokinase |
| chr11_-_94601862 | 3.27 |
ENSMUST00000103164.3
|
Acsf2
|
acyl-CoA synthetase family member 2 |
| chr9_+_107576915 | 3.26 |
ENSMUST00000112387.2
ENSMUST00000123005.1 ENSMUST00000010195.7 ENSMUST00000144392.1 |
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr3_+_94693556 | 3.00 |
ENSMUST00000090848.3
ENSMUST00000173981.1 ENSMUST00000173849.1 ENSMUST00000174223.1 |
Selenbp2
|
selenium binding protein 2 |
| chr19_-_4042165 | 2.90 |
ENSMUST00000042700.9
|
Gstp2
|
glutathione S-transferase, pi 2 |
| chr10_-_41579207 | 2.85 |
ENSMUST00000095227.3
|
Ccdc162
|
coiled-coil domain containing 162 |
| chr17_-_12507704 | 2.85 |
ENSMUST00000024595.2
|
Slc22a3
|
solute carrier family 22 (organic cation transporter), member 3 |
| chr17_+_8165501 | 2.80 |
ENSMUST00000097419.3
ENSMUST00000024636.8 |
Fgfr1op
|
Fgfr1 oncogene partner |
| chr5_+_45493374 | 2.79 |
ENSMUST00000046122.6
|
Lap3
|
leucine aminopeptidase 3 |
| chr13_-_29984219 | 2.73 |
ENSMUST00000146092.1
|
E2f3
|
E2F transcription factor 3 |
| chr3_-_116712198 | 2.73 |
ENSMUST00000120120.1
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr1_-_136260873 | 2.72 |
ENSMUST00000086395.5
|
Gpr25
|
G protein-coupled receptor 25 |
| chr16_-_97922582 | 2.72 |
ENSMUST00000170757.1
|
C2cd2
|
C2 calcium-dependent domain containing 2 |
| chr19_-_59076069 | 2.69 |
ENSMUST00000047511.7
ENSMUST00000163821.1 |
4930506M07Rik
|
RIKEN cDNA 4930506M07 gene |
| chr5_+_135106881 | 2.68 |
ENSMUST00000005507.3
|
Mlxipl
|
MLX interacting protein-like |
| chr12_-_57546121 | 2.53 |
ENSMUST00000044380.6
|
Foxa1
|
forkhead box A1 |
| chr17_-_57228003 | 2.49 |
ENSMUST00000177046.1
ENSMUST00000024988.8 |
C3
|
complement component 3 |
| chr2_+_69135799 | 2.42 |
ENSMUST00000041865.7
|
Nostrin
|
nitric oxide synthase trafficker |
| chr12_-_98577940 | 2.38 |
ENSMUST00000110113.1
|
Kcnk10
|
potassium channel, subfamily K, member 10 |
| chr2_+_31887262 | 2.38 |
ENSMUST00000138325.1
ENSMUST00000028187.6 |
Lamc3
|
laminin gamma 3 |
| chr3_-_116711820 | 2.37 |
ENSMUST00000153108.1
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr7_+_140845562 | 2.32 |
ENSMUST00000035300.5
|
Scgb1c1
|
secretoglobin, family 1C, member 1 |
| chr11_-_116199040 | 2.25 |
ENSMUST00000066587.5
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr6_+_4600840 | 2.24 |
ENSMUST00000015333.5
|
Casd1
|
CAS1 domain containing 1 |
| chrX_-_166479633 | 2.22 |
ENSMUST00000049435.8
|
Rab9
|
RAB9, member RAS oncogene family |
| chr11_-_50931612 | 2.18 |
ENSMUST00000109124.3
|
Zfp354b
|
zinc finger protein 354B |
| chr10_+_107271827 | 2.17 |
ENSMUST00000020057.8
ENSMUST00000105280.3 |
Lin7a
|
lin-7 homolog A (C. elegans) |
| chr1_-_51915901 | 2.16 |
ENSMUST00000018561.7
ENSMUST00000114537.2 |
Myo1b
|
myosin IB |
| chr11_+_57801575 | 2.12 |
ENSMUST00000020826.5
|
Sap30l
|
SAP30-like |
| chr12_-_103958939 | 2.10 |
ENSMUST00000122229.1
|
Serpina1e
|
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
| chr11_-_3863895 | 2.05 |
ENSMUST00000070552.7
|
Osbp2
|
oxysterol binding protein 2 |
| chr6_+_4601124 | 1.99 |
ENSMUST00000181734.1
ENSMUST00000141359.1 |
Casd1
|
CAS1 domain containing 1 |
| chr16_-_18426372 | 1.98 |
ENSMUST00000000335.4
|
Comt
|
catechol-O-methyltransferase |
| chr1_-_51915968 | 1.91 |
ENSMUST00000046390.7
|
Myo1b
|
myosin IB |
| chr5_+_65107551 | 1.91 |
ENSMUST00000101192.2
|
Klhl5
|
kelch-like 5 |
| chr8_+_86624043 | 1.86 |
ENSMUST00000034141.9
ENSMUST00000122188.1 |
Lonp2
|
lon peptidase 2, peroxisomal |
| chr16_-_10543028 | 1.85 |
ENSMUST00000184863.1
ENSMUST00000038281.5 |
Dexi
|
dexamethasone-induced transcript |
| chr13_-_54055650 | 1.85 |
ENSMUST00000021932.5
|
Drd1a
|
dopamine receptor D1A |
| chr19_+_31868754 | 1.83 |
ENSMUST00000075838.5
|
A1cf
|
APOBEC1 complementation factor |
| chr11_+_101468164 | 1.82 |
ENSMUST00000001347.6
|
Rnd2
|
Rho family GTPase 2 |
| chr8_-_25016743 | 1.81 |
ENSMUST00000084032.5
|
Adam9
|
a disintegrin and metallopeptidase domain 9 (meltrin gamma) |
| chr1_+_9547948 | 1.81 |
ENSMUST00000144177.1
|
Adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr7_+_143475094 | 1.80 |
ENSMUST00000105917.2
|
Slc22a18
|
solute carrier family 22 (organic cation transporter), member 18 |
| chr3_-_104220103 | 1.80 |
ENSMUST00000121198.1
ENSMUST00000122303.1 |
Magi3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr3_-_104220360 | 1.79 |
ENSMUST00000064371.7
|
Magi3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr1_+_156035392 | 1.78 |
ENSMUST00000111757.3
|
Tor1aip2
|
torsin A interacting protein 2 |
| chr17_+_6106464 | 1.76 |
ENSMUST00000142030.1
|
Tulp4
|
tubby like protein 4 |
| chr13_-_66932904 | 1.75 |
ENSMUST00000172597.1
|
Mterfd1
|
MTERF domain containing 1 |
| chr1_-_156035891 | 1.71 |
ENSMUST00000126448.1
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr2_-_130424242 | 1.70 |
ENSMUST00000089581.4
|
Pced1a
|
PC-esterase domain containing 1A |
| chr1_+_182124737 | 1.69 |
ENSMUST00000111018.1
ENSMUST00000027792.5 |
Srp9
|
signal recognition particle 9 |
| chr17_-_24696147 | 1.69 |
ENSMUST00000046839.8
|
Gfer
|
growth factor, erv1 (S. cerevisiae)-like (augmenter of liver regeneration) |
| chr7_+_28180272 | 1.68 |
ENSMUST00000173223.1
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr4_+_129287614 | 1.67 |
ENSMUST00000102599.3
|
Sync
|
syncoilin |
| chr8_+_45975514 | 1.66 |
ENSMUST00000034051.6
|
Ufsp2
|
UFM1-specific peptidase 2 |
| chr18_+_9212856 | 1.66 |
ENSMUST00000041080.5
|
Fzd8
|
frizzled homolog 8 (Drosophila) |
| chr7_+_28180226 | 1.66 |
ENSMUST00000172467.1
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr9_-_50693799 | 1.64 |
ENSMUST00000120622.1
|
Dixdc1
|
DIX domain containing 1 |
| chr13_-_66933080 | 1.62 |
ENSMUST00000021991.4
|
Mterfd1
|
MTERF domain containing 1 |
| chr13_-_34652671 | 1.62 |
ENSMUST00000053459.7
|
Pxdc1
|
PX domain containing 1 |
| chr4_-_103215147 | 1.61 |
ENSMUST00000150285.1
|
Slc35d1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
| chr1_+_128244122 | 1.60 |
ENSMUST00000027592.3
|
Ubxn4
|
UBX domain protein 4 |
| chr17_+_6106880 | 1.59 |
ENSMUST00000149756.1
|
Tulp4
|
tubby like protein 4 |
| chr10_+_43901782 | 1.59 |
ENSMUST00000054418.5
|
Rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr19_-_21652779 | 1.55 |
ENSMUST00000179768.1
ENSMUST00000178523.1 ENSMUST00000038830.3 |
1110059E24Rik
|
RIKEN cDNA 1110059E24 gene |
| chr5_-_45857473 | 1.55 |
ENSMUST00000016026.7
ENSMUST00000067997.6 ENSMUST00000045586.6 |
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chrX_-_44790146 | 1.55 |
ENSMUST00000115056.1
|
Dcaf12l1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr2_+_30364262 | 1.53 |
ENSMUST00000142801.1
ENSMUST00000100214.3 |
Fam73b
|
family with sequence similarity 73, member B |
| chr4_+_122995944 | 1.53 |
ENSMUST00000106252.2
|
Mycl
|
v-myc myelocytomatosis viral oncogene homolog, lung carcinoma derived (avian) |
| chr4_-_141825997 | 1.52 |
ENSMUST00000102481.3
|
Cela2a
|
chymotrypsin-like elastase family, member 2A |
| chr1_+_82586942 | 1.52 |
ENSMUST00000113457.2
|
Col4a3
|
collagen, type IV, alpha 3 |
| chrX_-_44790179 | 1.52 |
ENSMUST00000060481.2
|
Dcaf12l1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr6_+_21986887 | 1.52 |
ENSMUST00000151315.1
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr4_+_100095791 | 1.51 |
ENSMUST00000039630.5
|
Ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr8_-_79294928 | 1.49 |
ENSMUST00000048718.2
|
Mmaa
|
methylmalonic aciduria (cobalamin deficiency) type A |
| chr10_+_20148920 | 1.47 |
ENSMUST00000116259.3
|
Map7
|
microtubule-associated protein 7 |
| chr3_-_85746266 | 1.46 |
ENSMUST00000118408.1
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr1_+_38987806 | 1.45 |
ENSMUST00000027247.5
|
Pdcl3
|
phosducin-like 3 |
| chr9_+_107888129 | 1.45 |
ENSMUST00000035202.2
|
Mon1a
|
MON1 homolog A (yeast) |
| chr1_+_156035705 | 1.42 |
ENSMUST00000111754.2
ENSMUST00000133152.1 |
Tor1aip2
|
torsin A interacting protein 2 |
| chr4_-_114908892 | 1.41 |
ENSMUST00000068654.3
|
Foxd2
|
forkhead box D2 |
| chr15_-_77533312 | 1.40 |
ENSMUST00000062562.5
|
Apol7c
|
apolipoprotein L 7c |
| chr15_-_89196457 | 1.38 |
ENSMUST00000078953.7
|
Dennd6b
|
DENN/MADD domain containing 6B |
| chrX_-_85776606 | 1.36 |
ENSMUST00000142152.1
ENSMUST00000156390.1 ENSMUST00000113978.2 |
Gyk
|
glycerol kinase |
| chr7_-_80403315 | 1.36 |
ENSMUST00000147150.1
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr1_-_82586781 | 1.34 |
ENSMUST00000087050.5
|
Col4a4
|
collagen, type IV, alpha 4 |
| chr19_+_11770415 | 1.34 |
ENSMUST00000167199.1
|
Mrpl16
|
mitochondrial ribosomal protein L16 |
| chr7_-_19149189 | 1.32 |
ENSMUST00000032566.1
|
Qpctl
|
glutaminyl-peptide cyclotransferase-like |
| chr17_+_47436731 | 1.32 |
ENSMUST00000150819.2
|
AI661453
|
expressed sequence AI661453 |
| chr5_-_135744206 | 1.31 |
ENSMUST00000153399.1
ENSMUST00000043378.2 |
Tmem120a
|
transmembrane protein 120A |
| chr4_+_122996035 | 1.31 |
ENSMUST00000030407.7
|
Mycl
|
v-myc myelocytomatosis viral oncogene homolog, lung carcinoma derived (avian) |
| chr8_+_85060055 | 1.30 |
ENSMUST00000095220.3
|
Fbxw9
|
F-box and WD-40 domain protein 9 |
| chr10_-_30618436 | 1.29 |
ENSMUST00000161074.1
|
Hint3
|
histidine triad nucleotide binding protein 3 |
| chr2_+_126707319 | 1.29 |
ENSMUST00000028841.7
ENSMUST00000110416.2 |
Usp8
|
ubiquitin specific peptidase 8 |
| chr5_-_114443993 | 1.29 |
ENSMUST00000112245.1
|
Mmab
|
methylmalonic aciduria (cobalamin deficiency) type B homolog (human) |
| chr12_+_76533540 | 1.28 |
ENSMUST00000075249.4
|
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr7_-_45459839 | 1.28 |
ENSMUST00000094434.4
|
Ftl1
|
ferritin light chain 1 |
| chr11_-_21572193 | 1.28 |
ENSMUST00000102874.4
|
Mdh1
|
malate dehydrogenase 1, NAD (soluble) |
| chr3_-_79628859 | 1.27 |
ENSMUST00000029386.7
|
Etfdh
|
electron transferring flavoprotein, dehydrogenase |
| chr5_-_114444036 | 1.27 |
ENSMUST00000031560.7
|
Mmab
|
methylmalonic aciduria (cobalamin deficiency) type B homolog (human) |
| chr5_+_138255608 | 1.26 |
ENSMUST00000062067.6
|
Lamtor4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr16_+_5007283 | 1.26 |
ENSMUST00000184439.1
|
Smim22
|
small integral membrane protein 22 |
| chr7_-_127935429 | 1.26 |
ENSMUST00000141385.1
ENSMUST00000156152.1 |
Prss36
|
protease, serine, 36 |
| chr10_-_30618337 | 1.26 |
ENSMUST00000019925.5
|
Hint3
|
histidine triad nucleotide binding protein 3 |
| chr5_-_107869153 | 1.22 |
ENSMUST00000128723.1
ENSMUST00000124034.1 |
Evi5
|
ecotropic viral integration site 5 |
| chr4_-_124850652 | 1.22 |
ENSMUST00000125776.1
|
1110065P20Rik
|
RIKEN cDNA 1110065P20 gene |
| chr19_-_6921753 | 1.22 |
ENSMUST00000173635.1
|
Esrra
|
estrogen related receptor, alpha |
| chr5_+_121660869 | 1.21 |
ENSMUST00000111765.1
|
Brap
|
BRCA1 associated protein |
| chr2_+_104027823 | 1.21 |
ENSMUST00000111135.1
ENSMUST00000111136.1 ENSMUST00000102565.3 |
Fbxo3
|
F-box protein 3 |
| chr8_-_54529951 | 1.20 |
ENSMUST00000067476.8
|
Spcs3
|
signal peptidase complex subunit 3 homolog (S. cerevisiae) |
| chr7_+_4877047 | 1.19 |
ENSMUST00000125249.1
|
Isoc2a
|
isochorismatase domain containing 2a |
| chr11_-_115297510 | 1.19 |
ENSMUST00000056153.7
|
Fads6
|
fatty acid desaturase domain family, member 6 |
| chr2_+_127270208 | 1.19 |
ENSMUST00000110375.2
|
Stard7
|
START domain containing 7 |
| chr5_+_144255223 | 1.18 |
ENSMUST00000056578.6
|
Bri3
|
brain protein I3 |
| chr4_+_116558056 | 1.17 |
ENSMUST00000106475.1
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr2_-_130424673 | 1.16 |
ENSMUST00000110277.1
|
Pced1a
|
PC-esterase domain containing 1A |
| chr11_-_120796369 | 1.15 |
ENSMUST00000143139.1
ENSMUST00000129955.1 ENSMUST00000026151.4 ENSMUST00000167023.1 ENSMUST00000106133.1 ENSMUST00000106135.1 |
Dus1l
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr3_+_54755574 | 1.15 |
ENSMUST00000029371.2
|
Smad9
|
SMAD family member 9 |
| chrX_+_42067876 | 1.15 |
ENSMUST00000126375.1
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr18_+_84088077 | 1.14 |
ENSMUST00000060223.2
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr13_+_66932802 | 1.14 |
ENSMUST00000021990.3
|
Ptdss1
|
phosphatidylserine synthase 1 |
| chr12_-_81532840 | 1.13 |
ENSMUST00000169158.1
ENSMUST00000164431.1 ENSMUST00000163402.1 ENSMUST00000166664.1 ENSMUST00000164386.1 |
Synj2bp
Gm20498
|
synaptojanin 2 binding protein predicted gene 20498 |
| chr2_-_65022740 | 1.13 |
ENSMUST00000028252.7
|
Grb14
|
growth factor receptor bound protein 14 |
| chr2_-_160872985 | 1.13 |
ENSMUST00000109460.1
ENSMUST00000127201.1 |
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr5_+_108694222 | 1.13 |
ENSMUST00000013633.8
ENSMUST00000112560.3 |
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr8_+_53511695 | 1.13 |
ENSMUST00000033920.4
|
Aga
|
aspartylglucosaminidase |
| chr4_+_116557658 | 1.12 |
ENSMUST00000030460.8
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr1_+_44147847 | 1.11 |
ENSMUST00000027214.3
|
Ercc5
|
excision repair cross-complementing rodent repair deficiency, complementation group 5 |
| chr5_+_137569851 | 1.11 |
ENSMUST00000031729.8
|
Tfr2
|
transferrin receptor 2 |
| chr4_-_135385645 | 1.09 |
ENSMUST00000105857.1
ENSMUST00000105858.1 ENSMUST00000064481.8 ENSMUST00000123632.1 |
Ncmap
|
noncompact myelin associated protein |
| chr4_-_124850670 | 1.09 |
ENSMUST00000163946.1
ENSMUST00000106190.3 |
1110065P20Rik
|
RIKEN cDNA 1110065P20 gene |
| chr14_-_57398483 | 1.09 |
ENSMUST00000022517.7
|
Cryl1
|
crystallin, lambda 1 |
| chr2_+_113327711 | 1.09 |
ENSMUST00000099576.2
|
Fmn1
|
formin 1 |
| chr19_-_6909599 | 1.08 |
ENSMUST00000173091.1
|
Prdx5
|
peroxiredoxin 5 |
| chr3_-_116712644 | 1.08 |
ENSMUST00000029569.2
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr15_+_89076124 | 1.08 |
ENSMUST00000165690.1
|
Trabd
|
TraB domain containing |
| chr9_-_105495130 | 1.08 |
ENSMUST00000038118.7
|
Atp2c1
|
ATPase, Ca++-sequestering |
| chrX_+_100774741 | 1.07 |
ENSMUST00000113735.2
|
Dlg3
|
discs, large homolog 3 (Drosophila) |
| chr10_+_93641041 | 1.05 |
ENSMUST00000020204.4
|
Ntn4
|
netrin 4 |
| chr2_-_174438996 | 1.04 |
ENSMUST00000016400.8
|
Ctsz
|
cathepsin Z |
| chr2_-_160872829 | 1.03 |
ENSMUST00000176141.1
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr19_+_21653302 | 1.03 |
ENSMUST00000052556.3
|
Abhd17b
|
abhydrolase domain containing 17B |
| chr1_-_36709904 | 1.03 |
ENSMUST00000043951.3
|
Actr1b
|
ARP1 actin-related protein 1B, centractin beta |
| chr19_-_6941428 | 1.02 |
ENSMUST00000025909.4
ENSMUST00000099774.3 |
Gpr137
|
G protein-coupled receptor 137 |
| chr16_+_5007306 | 1.02 |
ENSMUST00000178155.2
ENSMUST00000184256.1 ENSMUST00000185147.1 |
Smim22
|
small integral membrane protein 22 |
| chrX_+_161717055 | 1.02 |
ENSMUST00000112338.1
|
Rai2
|
retinoic acid induced 2 |
| chr4_-_124850473 | 1.01 |
ENSMUST00000137769.2
|
1110065P20Rik
|
RIKEN cDNA 1110065P20 gene |
| chr13_+_93771656 | 1.00 |
ENSMUST00000091403.4
|
Arsb
|
arylsulfatase B |
| chr17_+_13760502 | 1.00 |
ENSMUST00000139347.1
ENSMUST00000156591.1 ENSMUST00000170827.2 ENSMUST00000139666.1 ENSMUST00000137784.1 ENSMUST00000137708.1 ENSMUST00000150848.1 |
Mllt4
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4 |
| chr16_-_11909398 | 0.99 |
ENSMUST00000127972.1
ENSMUST00000121750.1 ENSMUST00000096272.4 ENSMUST00000073371.6 |
Cpped1
|
calcineurin-like phosphoesterase domain containing 1 |
| chr18_-_35655185 | 0.99 |
ENSMUST00000097619.1
|
Prob1
|
proline rich basic protein 1 |
| chr17_-_26244118 | 0.99 |
ENSMUST00000118487.1
|
Itfg3
|
integrin alpha FG-GAP repeat containing 3 |
| chr11_-_3722189 | 0.98 |
ENSMUST00000102950.3
ENSMUST00000101632.3 |
Osbp2
|
oxysterol binding protein 2 |
| chr9_+_75441518 | 0.98 |
ENSMUST00000048937.4
|
Leo1
|
Leo1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) |
| chr6_+_59208870 | 0.98 |
ENSMUST00000062626.3
|
Tigd2
|
tigger transposable element derived 2 |
| chr15_+_94543666 | 0.97 |
ENSMUST00000109248.1
|
Irak4
|
interleukin-1 receptor-associated kinase 4 |
| chr3_-_89773221 | 0.97 |
ENSMUST00000038450.1
|
4632404H12Rik
|
RIKEN cDNA 4632404H12 gene |
| chr5_-_137684665 | 0.97 |
ENSMUST00000100544.4
ENSMUST00000031736.9 ENSMUST00000151839.1 |
Agfg2
|
ArfGAP with FG repeats 2 |
| chr4_+_138454305 | 0.96 |
ENSMUST00000050918.3
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr19_-_21652714 | 0.96 |
ENSMUST00000177577.1
|
1110059E24Rik
|
RIKEN cDNA 1110059E24 gene |
| chr17_-_26244203 | 0.96 |
ENSMUST00000114988.1
|
Itfg3
|
integrin alpha FG-GAP repeat containing 3 |
| chr7_+_44896125 | 0.95 |
ENSMUST00000166552.1
ENSMUST00000168207.1 |
Fuz
|
fuzzy homolog (Drosophila) |
| chr4_+_24898080 | 0.95 |
ENSMUST00000029925.3
ENSMUST00000151249.1 |
Ndufaf4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4 |
| chr3_+_132684106 | 0.94 |
ENSMUST00000169172.1
ENSMUST00000029664.6 |
Tbck
|
TBC1 domain containing kinase |
| chr8_-_95142477 | 0.94 |
ENSMUST00000034240.7
ENSMUST00000169748.1 |
Kifc3
|
kinesin family member C3 |
| chr7_+_48959089 | 0.94 |
ENSMUST00000183659.1
|
Nav2
|
neuron navigator 2 |
| chr5_+_121660528 | 0.93 |
ENSMUST00000031414.8
|
Brap
|
BRCA1 associated protein |
| chr17_-_25785533 | 0.93 |
ENSMUST00000140738.1
ENSMUST00000145053.1 ENSMUST00000138759.1 ENSMUST00000133071.1 ENSMUST00000077938.3 |
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chr13_+_80886095 | 0.93 |
ENSMUST00000161441.1
|
Arrdc3
|
arrestin domain containing 3 |
| chr9_+_67759423 | 0.92 |
ENSMUST00000171652.1
|
C2cd4b
|
C2 calcium-dependent domain containing 4B |
| chr15_-_37791993 | 0.90 |
ENSMUST00000168992.1
ENSMUST00000148652.1 |
Ncald
|
neurocalcin delta |
| chr13_+_67813740 | 0.90 |
ENSMUST00000181391.1
ENSMUST00000012725.7 |
Zfp273
|
zinc finger protein 273 |
| chr5_-_121618865 | 0.90 |
ENSMUST00000041252.6
ENSMUST00000111776.1 |
Acad12
|
acyl-Coenzyme A dehydrogenase family, member 12 |
| chr2_+_144556229 | 0.89 |
ENSMUST00000143573.1
ENSMUST00000028916.8 ENSMUST00000155258.1 |
Sec23b
|
SEC23B (S. cerevisiae) |
| chrX_-_8132770 | 0.89 |
ENSMUST00000130832.1
ENSMUST00000033506.6 ENSMUST00000115623.1 ENSMUST00000153839.1 |
Wdr13
|
WD repeat domain 13 |
| chr17_-_35909626 | 0.88 |
ENSMUST00000141132.1
|
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr12_-_24680890 | 0.88 |
ENSMUST00000156453.2
|
Cys1
|
cystin 1 |
| chr9_-_105495037 | 0.87 |
ENSMUST00000176190.1
ENSMUST00000163879.2 ENSMUST00000112558.2 ENSMUST00000176390.1 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr9_+_123150941 | 0.87 |
ENSMUST00000026890.4
|
Clec3b
|
C-type lectin domain family 3, member b |
| chr10_-_93081596 | 0.84 |
ENSMUST00000168617.1
ENSMUST00000168110.1 ENSMUST00000020200.7 |
Gm872
|
RIKEN cDNA 4930485B16 gene |
| chr10_+_86779000 | 0.84 |
ENSMUST00000099396.2
|
Nt5dc3
|
5'-nucleotidase domain containing 3 |
| chr9_-_44965519 | 0.83 |
ENSMUST00000125642.1
ENSMUST00000117506.1 ENSMUST00000117549.1 |
Ube4a
|
ubiquitination factor E4A, UFD2 homolog (S. cerevisiae) |
| chr5_+_93044720 | 0.83 |
ENSMUST00000180541.1
|
9330159N05Rik
|
RIKEN cDNA 9330159N05 gene |
| chr14_+_21500879 | 0.83 |
ENSMUST00000182964.1
|
Kat6b
|
K(lysine) acetyltransferase 6B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 15.0 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 1.2 | 6.2 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 1.1 | 3.3 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 1.1 | 3.3 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.9 | 6.5 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.8 | 5.9 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.8 | 2.5 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.8 | 11.5 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.8 | 17.7 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.7 | 3.3 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.6 | 2.5 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.6 | 1.9 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) |
| 0.6 | 1.9 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.5 | 2.5 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.5 | 1.9 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.5 | 3.7 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.5 | 1.8 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.4 | 1.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.4 | 2.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.4 | 1.2 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.4 | 2.7 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.4 | 1.1 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.4 | 2.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.4 | 1.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.4 | 2.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.4 | 2.2 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.3 | 1.3 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.3 | 2.2 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.3 | 1.3 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.3 | 0.9 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.3 | 2.7 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 1.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 1.7 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.3 | 1.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.3 | 0.8 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.3 | 0.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.3 | 0.8 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.2 | 3.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.2 | 0.7 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.2 | 2.8 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 0.7 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.2 | 3.9 | GO:0051608 | histamine transport(GO:0051608) |
| 0.2 | 1.3 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.2 | 2.9 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 0.4 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.2 | 0.6 | GO:0007172 | signal complex assembly(GO:0007172) regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.2 | 1.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 1.5 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.2 | 0.6 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.2 | 0.6 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.2 | 1.6 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.2 | 0.8 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.2 | 0.8 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 0.8 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.2 | 1.7 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.2 | 0.9 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 1.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 0.4 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.2 | 0.7 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.2 | 4.8 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.2 | 1.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 1.7 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 1.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.2 | 2.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 0.6 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.2 | 0.9 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 0.8 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 2.0 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.2 | 0.6 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.2 | 0.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 1.0 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.4 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.4 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 0.6 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.7 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.4 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 1.1 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.1 | 0.4 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 1.6 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 1.9 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 3.1 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 4.0 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 2.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.7 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.7 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.1 | 0.9 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 3.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.5 | GO:0055099 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.6 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.1 | 0.7 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.1 | 0.5 | GO:1900175 | regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.8 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.5 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.1 | 0.7 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.1 | 0.9 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 1.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.4 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.4 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.1 | 1.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.3 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 0.6 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 0.3 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.7 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.1 | 3.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.5 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.6 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.1 | 0.3 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.1 | 0.3 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.5 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.7 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.1 | 3.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.8 | GO:1903350 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.1 | 0.8 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 1.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 | 4.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.7 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.6 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 0.5 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 3.0 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 3.5 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.9 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 1.0 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.1 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 1.3 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 3.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.4 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 1.5 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 1.3 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.2 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.6 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 4.4 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.6 | GO:0009169 | ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 1.7 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 1.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0034182 | regulation of maintenance of sister chromatid cohesion(GO:0034091) positive regulation of maintenance of sister chromatid cohesion(GO:0034093) regulation of maintenance of mitotic sister chromatid cohesion(GO:0034182) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.6 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.7 | GO:0036119 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 1.8 | GO:0015893 | drug transport(GO:0015893) |
| 0.0 | 0.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.3 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 2.1 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 2.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.2 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.7 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 1.2 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.2 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.6 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.2 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 0.6 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.7 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 1.4 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 1.6 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 2.4 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.2 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 1.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.4 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 1.0 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 1.8 | GO:0006818 | hydrogen transport(GO:0006818) |
| 0.0 | 0.8 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.3 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.1 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 2.3 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.7 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.3 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.0 | 0.0 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 1.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.3 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 15.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 1.0 | 2.9 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.5 | 2.9 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.4 | 1.7 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.4 | 1.5 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.3 | 2.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 1.8 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.3 | 1.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.3 | 0.8 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 1.5 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.2 | 2.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 1.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 4.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 1.7 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.2 | 1.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 3.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 0.8 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.2 | 6.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 1.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 1.1 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.2 | 1.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 3.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 4.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.8 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.6 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 1.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.8 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.9 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.6 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.1 | 0.5 | GO:0032444 | activin responsive factor complex(GO:0032444) SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 4.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.5 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.3 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.9 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 2.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 1.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.8 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 1.1 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 4.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 3.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.4 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 1.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 16.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.7 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.1 | GO:0044439 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 5.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.7 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 2.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 1.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 4.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0030663 | COPI vesicle coat(GO:0030126) COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 1.1 | 6.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.0 | 11.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.8 | 3.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.8 | 15.0 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.7 | 5.4 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.7 | 2.0 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.6 | 2.5 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.6 | 1.9 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.6 | 2.8 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.5 | 2.8 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.5 | 1.9 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.4 | 1.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.4 | 1.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.4 | 1.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.4 | 2.2 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.4 | 1.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.4 | 2.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.4 | 4.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.3 | 1.8 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.3 | 3.3 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.3 | 1.9 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 1.6 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.3 | 6.2 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.2 | 1.0 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.2 | 1.7 | GO:0008312 | signal recognition particle binding(GO:0005047) 7S RNA binding(GO:0008312) |
| 0.2 | 0.9 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.2 | 1.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.2 | 0.6 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.2 | 4.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 0.8 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.2 | 1.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.2 | 1.7 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.2 | 1.5 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 0.5 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.2 | 1.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 1.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 4.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 0.8 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.2 | 0.6 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.2 | 1.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 2.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.4 | GO:0070546 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.7 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 0.7 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.7 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.7 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.8 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.4 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 1.6 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.4 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.1 | 0.5 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.8 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 1.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.5 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 1.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 2.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.8 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 1.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.4 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.6 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.5 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.8 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 2.8 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.1 | 4.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.3 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.4 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 1.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 19.3 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.1 | 0.8 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 1.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.9 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 0.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 1.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.7 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.8 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 3.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.3 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.8 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 3.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.2 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 2.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 1.8 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 3.1 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.7 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 0.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 8.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 3.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.0 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 1.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.0 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.7 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 1.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.4 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 3.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.5 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.3 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 1.0 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.1 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 3.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 3.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.1 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 1.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.8 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.0 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 1.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 3.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 1.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 5.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 2.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 3.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 4.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 2.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 2.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 2.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 3.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 3.5 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 2.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 3.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 3.0 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 6.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 4.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 2.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 1.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 0.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 3.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 6.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 3.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 3.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 2.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.7 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.0 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.5 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 1.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.8 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 2.8 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.6 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 2.0 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |