Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Nfic_Nfib
Z-value: 1.25
Transcription factors associated with Nfic_Nfib
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
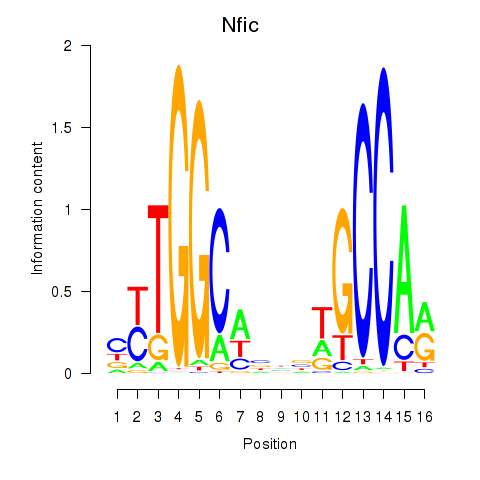
Nfic
|
ENSMUSG00000055053.11 | nuclear factor I/C |
|
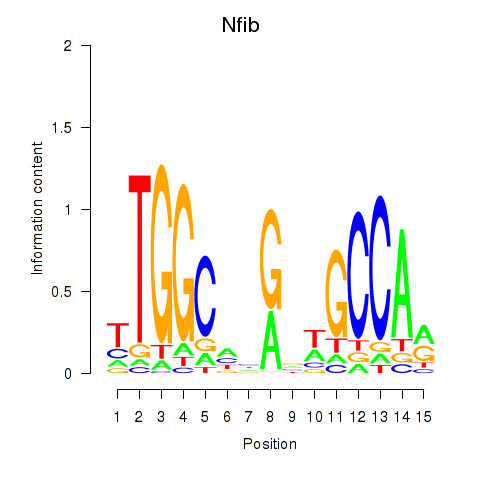
Nfib
|
ENSMUSG00000008575.11 | nuclear factor I/B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfib | mm10_v2_chr4_-_82705735_82705754 | 0.34 | 4.3e-02 | Click! |
| Nfic | mm10_v2_chr10_-_81427114_81427187 | 0.33 | 4.8e-02 | Click! |
Activity profile of Nfic_Nfib motif
Sorted Z-values of Nfic_Nfib motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_130369420 | 6.63 |
ENSMUST00000086029.3
|
Caln1
|
calneuron 1 |
| chr5_-_145805862 | 6.16 |
ENSMUST00000067479.5
|
Cyp3a44
|
cytochrome P450, family 3, subfamily a, polypeptide 44 |
| chr3_-_107221722 | 5.54 |
ENSMUST00000029504.8
|
Cym
|
chymosin |
| chr6_+_78425973 | 4.58 |
ENSMUST00000079926.5
|
Reg1
|
regenerating islet-derived 1 |
| chr7_+_119526269 | 3.98 |
ENSMUST00000066465.1
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr9_-_109849440 | 3.78 |
ENSMUST00000112022.2
|
Camp
|
cathelicidin antimicrobial peptide |
| chr11_+_115462464 | 3.57 |
ENSMUST00000106532.3
ENSMUST00000092445.5 ENSMUST00000153466.1 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr7_-_142576492 | 3.35 |
ENSMUST00000140716.1
|
H19
|
H19 fetal liver mRNA |
| chr7_-_44524642 | 3.18 |
ENSMUST00000165208.2
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr18_+_33464163 | 3.10 |
ENSMUST00000097634.3
|
Gm10549
|
predicted gene 10549 |
| chr17_-_32917320 | 3.08 |
ENSMUST00000179434.1
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr15_+_54745702 | 3.07 |
ENSMUST00000050027.8
|
Nov
|
nephroblastoma overexpressed gene |
| chr7_-_142578139 | 3.05 |
ENSMUST00000136359.1
|
H19
|
H19 fetal liver mRNA |
| chr17_-_46438471 | 3.04 |
ENSMUST00000087012.5
|
Slc22a7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chr17_-_12675833 | 2.95 |
ENSMUST00000024596.8
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr2_+_173153048 | 2.88 |
ENSMUST00000029017.5
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr7_-_142578093 | 2.81 |
ENSMUST00000149974.1
ENSMUST00000152754.1 |
H19
|
H19 fetal liver mRNA |
| chr11_+_61485431 | 2.63 |
ENSMUST00000064783.3
ENSMUST00000040522.6 |
Mfap4
|
microfibrillar-associated protein 4 |
| chr7_+_131032061 | 2.58 |
ENSMUST00000084509.3
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr5_-_145720124 | 2.50 |
ENSMUST00000094111.4
|
Cyp3a41a
|
cytochrome P450, family 3, subfamily a, polypeptide 41A |
| chr7_+_47050628 | 2.50 |
ENSMUST00000010451.5
|
Tmem86a
|
transmembrane protein 86A |
| chr8_+_94472763 | 2.43 |
ENSMUST00000053085.5
|
Nlrc5
|
NLR family, CARD domain containing 5 |
| chr17_-_32917048 | 2.37 |
ENSMUST00000054174.7
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr5_-_35525567 | 2.33 |
ENSMUST00000132959.1
|
Cpz
|
carboxypeptidase Z |
| chr4_+_130047914 | 2.33 |
ENSMUST00000142293.1
|
Col16a1
|
collagen, type XVI, alpha 1 |
| chr6_-_55175019 | 2.25 |
ENSMUST00000003569.5
|
Inmt
|
indolethylamine N-methyltransferase |
| chr5_-_134747241 | 2.23 |
ENSMUST00000015138.9
|
Eln
|
elastin |
| chr10_+_40629987 | 2.19 |
ENSMUST00000019977.7
|
Ddo
|
D-aspartate oxidase |
| chr11_+_109485606 | 2.16 |
ENSMUST00000106697.1
|
Arsg
|
arylsulfatase G |
| chr9_-_57683644 | 2.07 |
ENSMUST00000034860.3
|
Cyp1a2
|
cytochrome P450, family 1, subfamily a, polypeptide 2 |
| chr11_+_3989924 | 2.05 |
ENSMUST00000109981.1
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chrX_-_48034842 | 2.01 |
ENSMUST00000039026.7
|
Apln
|
apelin |
| chr6_-_83656082 | 1.90 |
ENSMUST00000014686.2
|
Clec4f
|
C-type lectin domain family 4, member f |
| chr6_-_115251839 | 1.90 |
ENSMUST00000032462.6
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr9_-_71163224 | 1.87 |
ENSMUST00000074465.2
|
Aqp9
|
aquaporin 9 |
| chr1_+_93006328 | 1.81 |
ENSMUST00000059676.4
|
Aqp12
|
aquaporin 12 |
| chr5_-_53707532 | 1.75 |
ENSMUST00000031093.3
|
Cckar
|
cholecystokinin A receptor |
| chrX_-_136068236 | 1.74 |
ENSMUST00000049130.7
|
Bex2
|
brain expressed X-linked 2 |
| chr3_-_8667033 | 1.70 |
ENSMUST00000042412.3
|
Hey1
|
hairy/enhancer-of-split related with YRPW motif 1 |
| chr2_+_25289899 | 1.68 |
ENSMUST00000028337.6
|
Lrrc26
|
leucine rich repeat containing 26 |
| chr18_-_33463747 | 1.67 |
ENSMUST00000171533.1
|
Nrep
|
neuronal regeneration related protein |
| chr12_+_110279228 | 1.63 |
ENSMUST00000097228.4
|
Dio3
|
deiodinase, iodothyronine type III |
| chrX_+_136270253 | 1.61 |
ENSMUST00000178632.1
ENSMUST00000053540.4 |
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr11_+_104550663 | 1.61 |
ENSMUST00000018800.2
|
Myl4
|
myosin, light polypeptide 4 |
| chr5_+_146079254 | 1.60 |
ENSMUST00000035571.8
|
Cyp3a59
|
cytochrome P450, family 3, subfamily a, polypeptide 59 |
| chrX_-_52613936 | 1.59 |
ENSMUST00000114857.1
|
Gpc3
|
glypican 3 |
| chrX_-_52613913 | 1.57 |
ENSMUST00000069360.7
|
Gpc3
|
glypican 3 |
| chr7_+_19411086 | 1.56 |
ENSMUST00000003643.1
|
Ckm
|
creatine kinase, muscle |
| chr17_-_28517509 | 1.53 |
ENSMUST00000114792.1
ENSMUST00000177939.1 |
Fkbp5
|
FK506 binding protein 5 |
| chrX_+_136270302 | 1.47 |
ENSMUST00000113112.1
|
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr9_+_46240696 | 1.45 |
ENSMUST00000034585.6
|
Apoa4
|
apolipoprotein A-IV |
| chr18_-_33464007 | 1.42 |
ENSMUST00000168890.1
|
Nrep
|
neuronal regeneration related protein |
| chr4_-_57916283 | 1.40 |
ENSMUST00000063816.5
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene |
| chr5_-_135078224 | 1.38 |
ENSMUST00000067935.4
ENSMUST00000076203.2 |
Vps37d
|
vacuolar protein sorting 37D (yeast) |
| chr11_-_110095937 | 1.38 |
ENSMUST00000106664.3
ENSMUST00000046223.7 ENSMUST00000106662.1 |
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr8_-_104631312 | 1.35 |
ENSMUST00000034351.6
|
Rrad
|
Ras-related associated with diabetes |
| chr13_+_43785107 | 1.35 |
ENSMUST00000015540.2
|
Cd83
|
CD83 antigen |
| chr2_+_27079371 | 1.35 |
ENSMUST00000091233.6
|
Adamtsl2
|
ADAMTS-like 2 |
| chr15_-_98296083 | 1.28 |
ENSMUST00000169721.1
ENSMUST00000023722.5 |
Zfp641
|
zinc finger protein 641 |
| chr11_+_115877497 | 1.28 |
ENSMUST00000144032.1
|
Myo15b
|
myosin XVB |
| chr14_-_54994541 | 1.26 |
ENSMUST00000153783.1
ENSMUST00000168485.1 ENSMUST00000102803.3 |
Myh7
|
myosin, heavy polypeptide 7, cardiac muscle, beta |
| chr19_+_6164433 | 1.26 |
ENSMUST00000045042.7
|
Batf2
|
basic leucine zipper transcription factor, ATF-like 2 |
| chr3_+_90669063 | 1.25 |
ENSMUST00000069927.8
|
S100a8
|
S100 calcium binding protein A8 (calgranulin A) |
| chr5_-_38480131 | 1.24 |
ENSMUST00000143758.1
ENSMUST00000067886.5 |
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr10_-_105574435 | 1.21 |
ENSMUST00000061506.8
|
Tmtc2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr1_-_150466165 | 1.19 |
ENSMUST00000162367.1
ENSMUST00000161611.1 ENSMUST00000161320.1 ENSMUST00000159035.1 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr11_-_109722214 | 1.18 |
ENSMUST00000020938.7
|
Fam20a
|
family with sequence similarity 20, member A |
| chr13_-_41847626 | 1.17 |
ENSMUST00000121404.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr13_-_41847599 | 1.17 |
ENSMUST00000179758.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr17_+_31296191 | 1.16 |
ENSMUST00000165149.1
|
Slc37a1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
| chr17_+_29318850 | 1.16 |
ENSMUST00000114701.2
|
Pi16
|
peptidase inhibitor 16 |
| chr9_-_50752348 | 1.15 |
ENSMUST00000042790.3
|
Hspb2
|
heat shock protein 2 |
| chr2_+_119167758 | 1.14 |
ENSMUST00000057454.3
|
Gchfr
|
GTP cyclohydrolase I feedback regulator |
| chr1_-_162859684 | 1.12 |
ENSMUST00000131058.1
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr5_-_139813237 | 1.11 |
ENSMUST00000110832.1
|
Tmem184a
|
transmembrane protein 184a |
| chr10_-_75932468 | 1.10 |
ENSMUST00000120281.1
ENSMUST00000000924.6 |
Mmp11
|
matrix metallopeptidase 11 |
| chr3_-_151749877 | 1.10 |
ENSMUST00000029671.7
|
Ifi44
|
interferon-induced protein 44 |
| chr11_-_58613481 | 1.10 |
ENSMUST00000048801.7
|
2210407C18Rik
|
RIKEN cDNA 2210407C18 gene |
| chr2_-_93849679 | 1.07 |
ENSMUST00000068513.4
ENSMUST00000041593.8 ENSMUST00000130077.1 |
Accs
|
1-aminocyclopropane-1-carboxylate synthase (non-functional) |
| chr5_-_35525691 | 1.06 |
ENSMUST00000038676.5
|
Cpz
|
carboxypeptidase Z |
| chr16_+_30008657 | 1.06 |
ENSMUST00000181485.1
|
4632428C04Rik
|
RIKEN cDNA 4632428C04 gene |
| chr18_-_33463615 | 1.05 |
ENSMUST00000051087.8
|
Nrep
|
neuronal regeneration related protein |
| chr13_-_41847482 | 1.04 |
ENSMUST00000072012.3
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr9_+_62677826 | 1.02 |
ENSMUST00000034774.8
|
Itga11
|
integrin alpha 11 |
| chr18_+_37806389 | 1.01 |
ENSMUST00000076807.4
|
Pcdhga9
|
protocadherin gamma subfamily A, 9 |
| chr2_-_32451396 | 0.99 |
ENSMUST00000028160.8
ENSMUST00000113310.2 |
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr6_-_118780324 | 0.98 |
ENSMUST00000112793.3
ENSMUST00000075591.6 ENSMUST00000078320.7 ENSMUST00000112790.2 |
Cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit |
| chr17_-_34743849 | 0.97 |
ENSMUST00000069507.8
|
C4b
|
complement component 4B (Chido blood group) |
| chr17_-_27728889 | 0.96 |
ENSMUST00000167489.1
ENSMUST00000138970.1 ENSMUST00000114870.1 ENSMUST00000025054.2 |
Spdef
|
SAM pointed domain containing ets transcription factor |
| chr2_+_13573927 | 0.95 |
ENSMUST00000141365.1
ENSMUST00000028062.2 |
Vim
|
vimentin |
| chr19_+_6084983 | 0.95 |
ENSMUST00000025704.2
|
Cdca5
|
cell division cycle associated 5 |
| chr2_-_110314525 | 0.95 |
ENSMUST00000133608.1
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
| chr9_+_44134562 | 0.95 |
ENSMUST00000034650.8
ENSMUST00000098852.2 |
Mcam
|
melanoma cell adhesion molecule |
| chr14_+_59625281 | 0.94 |
ENSMUST00000053949.5
|
Shisa2
|
shisa homolog 2 (Xenopus laevis) |
| chr3_-_104818224 | 0.94 |
ENSMUST00000002297.5
|
Mov10
|
Moloney leukemia virus 10 |
| chr17_+_32685655 | 0.93 |
ENSMUST00000008801.6
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr8_-_64693027 | 0.93 |
ENSMUST00000048967.7
|
Cpe
|
carboxypeptidase E |
| chr8_-_123894736 | 0.93 |
ENSMUST00000034453.4
|
Acta1
|
actin, alpha 1, skeletal muscle |
| chr3_-_131272077 | 0.93 |
ENSMUST00000029610.8
|
Hadh
|
hydroxyacyl-Coenzyme A dehydrogenase |
| chr1_+_87574016 | 0.92 |
ENSMUST00000166259.1
ENSMUST00000172222.1 ENSMUST00000163606.1 |
Neu2
|
neuraminidase 2 |
| chr1_+_172481788 | 0.92 |
ENSMUST00000127052.1
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr16_+_90831113 | 0.92 |
ENSMUST00000037539.7
ENSMUST00000099543.3 |
Eva1c
|
eva-1 homolog C (C. elegans) |
| chr4_+_141239499 | 0.92 |
ENSMUST00000141834.2
|
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr11_-_117873433 | 0.92 |
ENSMUST00000033230.7
|
Tha1
|
threonine aldolase 1 |
| chr2_-_164404606 | 0.91 |
ENSMUST00000109359.1
ENSMUST00000109358.1 ENSMUST00000103103.3 |
Matn4
|
matrilin 4 |
| chr2_-_32424005 | 0.90 |
ENSMUST00000113307.2
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr16_+_34784917 | 0.90 |
ENSMUST00000023538.8
|
Mylk
|
myosin, light polypeptide kinase |
| chr3_-_104818266 | 0.89 |
ENSMUST00000168015.1
|
Mov10
|
Moloney leukemia virus 10 |
| chr5_+_30013141 | 0.89 |
ENSMUST00000026845.7
|
Il6
|
interleukin 6 |
| chr5_+_135725713 | 0.89 |
ENSMUST00000127096.1
|
Por
|
P450 (cytochrome) oxidoreductase |
| chr4_-_43454563 | 0.89 |
ENSMUST00000107926.1
|
Cd72
|
CD72 antigen |
| chr9_-_121792478 | 0.87 |
ENSMUST00000035110.4
|
Hhatl
|
hedgehog acyltransferase-like |
| chr2_+_129592914 | 0.87 |
ENSMUST00000103203.1
|
Sirpa
|
signal-regulatory protein alpha |
| chr2_+_129592818 | 0.87 |
ENSMUST00000153491.1
ENSMUST00000161620.1 ENSMUST00000179001.1 |
Sirpa
|
signal-regulatory protein alpha |
| chrX_-_101734125 | 0.86 |
ENSMUST00000056614.6
|
Cxcr3
|
chemokine (C-X-C motif) receptor 3 |
| chr17_+_33524170 | 0.85 |
ENSMUST00000087623.6
|
Adamts10
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 10 |
| chrX_-_107403295 | 0.84 |
ENSMUST00000033591.5
|
Itm2a
|
integral membrane protein 2A |
| chr4_-_43454582 | 0.83 |
ENSMUST00000107925.1
|
Cd72
|
CD72 antigen |
| chr18_-_32139570 | 0.83 |
ENSMUST00000171765.1
|
Proc
|
protein C |
| chr1_+_43730593 | 0.83 |
ENSMUST00000027217.8
|
1500015O10Rik
|
RIKEN cDNA 1500015O10 gene |
| chr1_+_92973113 | 0.82 |
ENSMUST00000027489.7
|
Gpr35
|
G protein-coupled receptor 35 |
| chr6_+_29433248 | 0.82 |
ENSMUST00000101617.2
ENSMUST00000065090.5 |
Flnc
|
filamin C, gamma |
| chr11_+_67277124 | 0.82 |
ENSMUST00000019625.5
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr17_+_32621319 | 0.82 |
ENSMUST00000077639.5
|
Gm9705
|
predicted gene 9705 |
| chr11_-_110095974 | 0.81 |
ENSMUST00000100287.2
|
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr5_-_137921612 | 0.80 |
ENSMUST00000031741.7
|
Cyp3a13
|
cytochrome P450, family 3, subfamily a, polypeptide 13 |
| chr15_+_80671829 | 0.79 |
ENSMUST00000023044.5
|
Fam83f
|
family with sequence similarity 83, member F |
| chr8_+_84148252 | 0.77 |
ENSMUST00000093375.4
|
4930432K21Rik
|
RIKEN cDNA 4930432K21 gene |
| chr11_-_95699143 | 0.77 |
ENSMUST00000062249.2
|
Gm9796
|
predicted gene 9796 |
| chr11_+_115440540 | 0.77 |
ENSMUST00000093914.4
|
4933422H20Rik
|
RIKEN cDNA 4933422H20 gene |
| chr10_-_81291227 | 0.77 |
ENSMUST00000045744.6
|
Tjp3
|
tight junction protein 3 |
| chr14_-_70520254 | 0.76 |
ENSMUST00000022693.7
|
Bmp1
|
bone morphogenetic protein 1 |
| chr3_+_54755574 | 0.75 |
ENSMUST00000029371.2
|
Smad9
|
SMAD family member 9 |
| chrX_+_151046131 | 0.75 |
ENSMUST00000112685.1
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr10_-_77113676 | 0.73 |
ENSMUST00000072755.4
ENSMUST00000105409.1 |
Col18a1
|
collagen, type XVIII, alpha 1 |
| chr19_-_5924797 | 0.73 |
ENSMUST00000055458.4
|
Cdc42ep2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr4_+_48045144 | 0.73 |
ENSMUST00000030025.3
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr12_-_87444017 | 0.73 |
ENSMUST00000091090.4
|
2700073G19Rik
|
RIKEN cDNA 2700073G19 gene |
| chr2_-_164833438 | 0.72 |
ENSMUST00000042775.4
|
Neurl2
|
neuralized-like 2 (Drosophila) |
| chr4_+_133480126 | 0.72 |
ENSMUST00000051676.6
|
Fam46b
|
family with sequence similarity 46, member B |
| chr1_+_133365160 | 0.71 |
ENSMUST00000129213.1
|
Etnk2
|
ethanolamine kinase 2 |
| chr4_+_130047840 | 0.71 |
ENSMUST00000044565.8
ENSMUST00000132251.1 |
Col16a1
|
collagen, type XVI, alpha 1 |
| chr2_+_85136355 | 0.71 |
ENSMUST00000057019.7
|
Aplnr
|
apelin receptor |
| chr4_+_43727181 | 0.70 |
ENSMUST00000095109.3
|
Hrct1
|
histidine rich carboxyl terminus 1 |
| chr11_-_69685537 | 0.69 |
ENSMUST00000018896.7
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr8_-_12573311 | 0.69 |
ENSMUST00000180858.1
|
D630011A20Rik
|
RIKEN cDNA D630011A20 gene |
| chrX_-_150812932 | 0.69 |
ENSMUST00000131241.1
ENSMUST00000147152.1 ENSMUST00000143843.1 |
Maged2
|
melanoma antigen, family D, 2 |
| chr9_+_44773191 | 0.68 |
ENSMUST00000147559.1
|
Ift46
|
intraflagellar transport 46 |
| chr7_-_31051431 | 0.67 |
ENSMUST00000073892.4
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr15_+_89322969 | 0.67 |
ENSMUST00000066991.5
|
Adm2
|
adrenomedullin 2 |
| chr5_-_110286159 | 0.67 |
ENSMUST00000031472.5
|
Pxmp2
|
peroxisomal membrane protein 2 |
| chr7_-_105482197 | 0.67 |
ENSMUST00000047040.2
|
Prkcdbp
|
protein kinase C, delta binding protein |
| chr11_+_78512193 | 0.66 |
ENSMUST00000001127.4
|
Poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr7_-_45694369 | 0.66 |
ENSMUST00000040636.6
|
Sec1
|
secretory blood group 1 |
| chr10_+_128254131 | 0.66 |
ENSMUST00000060782.3
|
Apon
|
apolipoprotein N |
| chr7_+_130936172 | 0.66 |
ENSMUST00000006367.7
|
Htra1
|
HtrA serine peptidase 1 |
| chr7_-_118995211 | 0.65 |
ENSMUST00000008878.8
|
Gprc5b
|
G protein-coupled receptor, family C, group 5, member B |
| chr7_-_134232005 | 0.65 |
ENSMUST00000134504.1
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr2_-_103283760 | 0.65 |
ENSMUST00000111174.1
|
Ehf
|
ets homologous factor |
| chr2_-_29253001 | 0.65 |
ENSMUST00000071201.4
|
Ntng2
|
netrin G2 |
| chr17_+_34204080 | 0.64 |
ENSMUST00000138491.1
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr17_+_29319183 | 0.64 |
ENSMUST00000114699.1
ENSMUST00000155348.1 |
Pi16
|
peptidase inhibitor 16 |
| chr14_-_30008817 | 0.63 |
ENSMUST00000122205.1
ENSMUST00000016110.6 |
Il17rb
|
interleukin 17 receptor B |
| chr7_-_19770509 | 0.63 |
ENSMUST00000003061.7
|
Bcam
|
basal cell adhesion molecule |
| chr1_-_75278345 | 0.63 |
ENSMUST00000039534.4
|
Resp18
|
regulated endocrine-specific protein 18 |
| chr8_+_22757744 | 0.61 |
ENSMUST00000033941.5
|
Plat
|
plasminogen activator, tissue |
| chr7_+_29309429 | 0.61 |
ENSMUST00000137848.1
|
Dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr2_+_120629113 | 0.61 |
ENSMUST00000150912.1
ENSMUST00000180041.1 |
Stard9
|
START domain containing 9 |
| chr10_-_77113928 | 0.61 |
ENSMUST00000149744.1
|
Col18a1
|
collagen, type XVIII, alpha 1 |
| chr6_-_113501818 | 0.61 |
ENSMUST00000101059.1
|
Prrt3
|
proline-rich transmembrane protein 3 |
| chr15_-_103366763 | 0.60 |
ENSMUST00000023128.6
|
Itga5
|
integrin alpha 5 (fibronectin receptor alpha) |
| chr14_+_65970610 | 0.60 |
ENSMUST00000127387.1
|
Clu
|
clusterin |
| chr6_+_29433131 | 0.60 |
ENSMUST00000090474.4
|
Flnc
|
filamin C, gamma |
| chr9_+_44773027 | 0.60 |
ENSMUST00000125877.1
|
Ift46
|
intraflagellar transport 46 |
| chr19_+_32619997 | 0.59 |
ENSMUST00000025833.6
|
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr14_-_70175397 | 0.59 |
ENSMUST00000143393.1
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr4_-_155019399 | 0.58 |
ENSMUST00000126098.1
ENSMUST00000176194.1 |
Plch2
|
phospholipase C, eta 2 |
| chr3_-_89245159 | 0.58 |
ENSMUST00000090924.6
|
Trim46
|
tripartite motif-containing 46 |
| chr19_-_42752710 | 0.58 |
ENSMUST00000076505.3
|
Pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr9_+_69454066 | 0.58 |
ENSMUST00000134907.1
|
Anxa2
|
annexin A2 |
| chrX_-_150813637 | 0.57 |
ENSMUST00000112700.1
|
Maged2
|
melanoma antigen, family D, 2 |
| chr11_+_50602072 | 0.57 |
ENSMUST00000040523.8
|
Adamts2
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 2 |
| chr2_+_11705712 | 0.57 |
ENSMUST00000138856.1
ENSMUST00000078834.5 ENSMUST00000114834.3 ENSMUST00000114833.3 ENSMUST00000114831.2 ENSMUST00000114832.2 |
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr2_-_180225812 | 0.56 |
ENSMUST00000015791.5
|
Lama5
|
laminin, alpha 5 |
| chr10_-_40025253 | 0.55 |
ENSMUST00000163705.2
|
AI317395
|
expressed sequence AI317395 |
| chr15_+_31568791 | 0.55 |
ENSMUST00000162532.1
|
Cmbl
|
carboxymethylenebutenolidase-like (Pseudomonas) |
| chr5_+_149006948 | 0.55 |
ENSMUST00000124198.1
|
Gm15408
|
predicted gene 15408 |
| chr14_+_65970804 | 0.55 |
ENSMUST00000138191.1
|
Clu
|
clusterin |
| chr6_+_135362931 | 0.55 |
ENSMUST00000032330.9
|
Emp1
|
epithelial membrane protein 1 |
| chr15_-_89377039 | 0.55 |
ENSMUST00000023285.4
|
Tymp
|
thymidine phosphorylase |
| chr15_+_34306666 | 0.54 |
ENSMUST00000163455.2
ENSMUST00000022947.5 |
Matn2
|
matrilin 2 |
| chr1_-_90843916 | 0.54 |
ENSMUST00000130846.2
ENSMUST00000097653.4 ENSMUST00000056925.9 |
Col6a3
|
collagen, type VI, alpha 3 |
| chr2_-_26604267 | 0.54 |
ENSMUST00000028286.5
|
Agpat2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) |
| chr11_+_4236411 | 0.54 |
ENSMUST00000075221.2
|
Osm
|
oncostatin M |
| chr3_+_52268337 | 0.54 |
ENSMUST00000053764.5
|
Foxo1
|
forkhead box O1 |
| chr12_-_112673944 | 0.54 |
ENSMUST00000130342.1
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr1_-_72874877 | 0.54 |
ENSMUST00000027377.8
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr15_-_78204228 | 0.53 |
ENSMUST00000005860.9
|
Pvalb
|
parvalbumin |
| chr11_-_82764303 | 0.53 |
ENSMUST00000021040.3
ENSMUST00000100722.4 |
Cct6b
|
chaperonin containing Tcp1, subunit 6b (zeta) |
| chr12_-_78983476 | 0.53 |
ENSMUST00000070174.7
|
Tmem229b
|
transmembrane protein 229B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfic_Nfib
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 1.0 | 3.1 | GO:1990523 | negative regulation of SMAD protein import into nucleus(GO:0060392) endothelial cell-cell adhesion(GO:0071603) negative regulation of sensory perception of pain(GO:1904057) bone regeneration(GO:1990523) |
| 1.0 | 2.9 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.7 | 3.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.7 | 2.2 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.7 | 9.2 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.7 | 2.0 | GO:1904020 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.6 | 3.2 | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 0.6 | 1.7 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.6 | 2.3 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.5 | 1.5 | GO:0044240 | negative regulation of lipoprotein oxidation(GO:0034443) multicellular organism lipid catabolic process(GO:0044240) |
| 0.5 | 1.9 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.4 | 1.7 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.4 | 1.2 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 0.4 | 5.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.3 | 2.0 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.3 | 1.0 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.3 | 1.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 2.4 | GO:0045345 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.3 | 0.9 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.3 | 0.9 | GO:0050748 | negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.3 | 1.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.3 | 0.8 | GO:0002591 | positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) peptide antigen transport(GO:0046968) |
| 0.3 | 2.6 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.3 | 1.3 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.2 | 0.9 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 3.8 | GO:0051873 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.2 | 0.9 | GO:0030070 | insulin processing(GO:0030070) |
| 0.2 | 0.7 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.2 | 0.9 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.2 | 2.0 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 1.8 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.2 | 0.9 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.2 | 0.9 | GO:0043602 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.2 | 1.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 0.9 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 | 0.6 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.2 | 3.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 1.0 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.2 | 1.0 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.2 | 1.1 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.2 | 2.7 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.2 | 0.7 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) |
| 0.2 | 1.9 | GO:0051132 | NK T cell activation(GO:0051132) |
| 0.2 | 1.7 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.2 | 1.2 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.2 | 1.4 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.2 | 0.7 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.2 | 0.8 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 1.5 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 1.1 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.2 | 1.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.2 | 0.9 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 0.6 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.1 | 0.4 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.1 | 0.8 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 2.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.5 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.4 | GO:1904209 | chemokine (C-C motif) ligand 2 secretion(GO:0035926) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 1.9 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.8 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.7 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.6 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.5 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.8 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.1 | 0.9 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.8 | GO:0015675 | nickel cation transport(GO:0015675) |
| 0.1 | 0.7 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 | 0.4 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 0.3 | GO:0003195 | tricuspid valve formation(GO:0003195) |
| 0.1 | 0.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.5 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.8 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 2.8 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.8 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.5 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.1 | 3.1 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.3 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 0.3 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.5 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.3 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.1 | 0.3 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) histone H3-R17 methylation(GO:0034971) negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.1 | 0.4 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.4 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.1 | 0.6 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.1 | 0.3 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.4 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.1 | 0.2 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 | 2.4 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 1.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.7 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.1 | 0.7 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 0.3 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.4 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 0.3 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.1 | 0.1 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.1 | 0.2 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.1 | 0.6 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 | 0.2 | GO:0070429 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.1 | 1.0 | GO:0031269 | pseudopodium assembly(GO:0031269) |
| 0.1 | 0.9 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.1 | 0.1 | GO:2000349 | negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.1 | 4.5 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 1.3 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 1.7 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 1.4 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.1 | 1.1 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.2 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.1 | 0.5 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 1.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.9 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 2.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.2 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.8 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.1 | 0.9 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.2 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.4 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 1.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.3 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.5 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.0 | 0.5 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.4 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 1.0 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.3 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.2 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.9 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.5 | GO:0051969 | regulation of transmission of nerve impulse(GO:0051969) |
| 0.0 | 0.5 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.4 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 2.0 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.3 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.2 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.2 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 | 0.2 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.4 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.3 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 0.2 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.4 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.0 | 0.3 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 1.4 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 2.7 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.3 | GO:0009169 | ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.5 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.0 | 0.4 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 1.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.5 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.3 | GO:0071285 | response to lithium ion(GO:0010226) cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.6 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 1.5 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.7 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.0 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 2.4 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) |
| 0.0 | 1.5 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.8 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.3 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.4 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.4 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.7 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.5 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.5 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.2 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.3 | GO:0098969 | neurotransmitter receptor transport to plasma membrane(GO:0098877) neurotransmitter receptor transport to postsynaptic membrane(GO:0098969) establishment of protein localization to postsynaptic membrane(GO:1903540) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.7 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.3 | GO:2000778 | positive regulation of interleukin-6 secretion(GO:2000778) |
| 0.0 | 0.0 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.3 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.2 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:2001198 | regulation of dendritic cell differentiation(GO:2001198) negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 2.2 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.0 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.4 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 0.7 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.0 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.4 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.2 | GO:0071624 | positive regulation of granulocyte chemotaxis(GO:0071624) |
| 0.0 | 0.3 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.4 | 1.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.4 | 2.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.3 | 1.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.3 | 3.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 0.9 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 5.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 6.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 4.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 0.6 | GO:0060187 | cell pole(GO:0060187) |
| 0.2 | 1.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 2.0 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.7 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.6 | GO:0005610 | laminin-5 complex(GO:0005610) laminin-10 complex(GO:0043259) |
| 0.1 | 9.6 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.0 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 1.0 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 0.6 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 1.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.4 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.1 | 0.8 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) TAP complex(GO:0042825) |
| 0.1 | 1.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 2.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 4.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.5 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.4 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 0.8 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.6 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 1.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 5.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 1.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.1 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 9.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 4.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.0 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 3.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.0 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 3.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.7 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 1.3 | 10.3 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 1.0 | 3.0 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.6 | 4.0 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.6 | 1.7 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.5 | 2.2 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.5 | 1.6 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.4 | 1.7 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.4 | 5.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.4 | 2.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.4 | 1.9 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.3 | 2.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.3 | 1.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.3 | 1.3 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.3 | 0.9 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.3 | 2.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.3 | 1.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 0.9 | GO:0047726 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.2 | 1.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 0.7 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.2 | 1.7 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 5.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 1.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 1.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 1.5 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.2 | 0.9 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.2 | 0.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 3.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 0.8 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.2 | 0.8 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.2 | 1.9 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 1.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) RAGE receptor binding(GO:0050786) |
| 0.1 | 2.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 4.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.4 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.1 | 3.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 1.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 2.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.7 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 0.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 1.7 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.8 | GO:0015099 | nickel cation transmembrane transporter activity(GO:0015099) |
| 0.1 | 0.3 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 0.7 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.4 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 0.3 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.1 | 3.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.9 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.3 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 1.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.9 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.7 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 2.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 2.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 1.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.4 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 3.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 3.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 2.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.3 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 0.1 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.2 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.1 | 1.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 1.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 2.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 1.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 6.2 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 1.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 1.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.6 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.3 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 2.0 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 0.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0070737 | protein-glycine ligase activity, elongating(GO:0070737) |
| 0.0 | 0.7 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 1.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 1.1 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 2.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.7 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.3 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 2.2 | GO:0004386 | helicase activity(GO:0004386) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 5.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 4.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 0.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.8 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.9 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 8.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 2.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 6.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 5.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.5 | 6.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 3.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 2.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 1.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 3.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 6.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 3.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 0.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 3.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 2.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 4.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 0.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 2.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.0 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 2.9 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 2.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.9 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |