Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
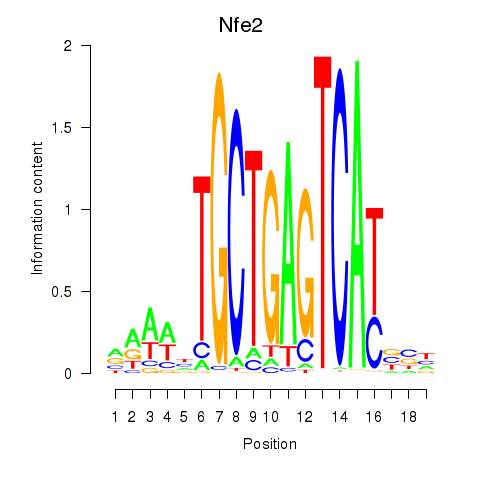
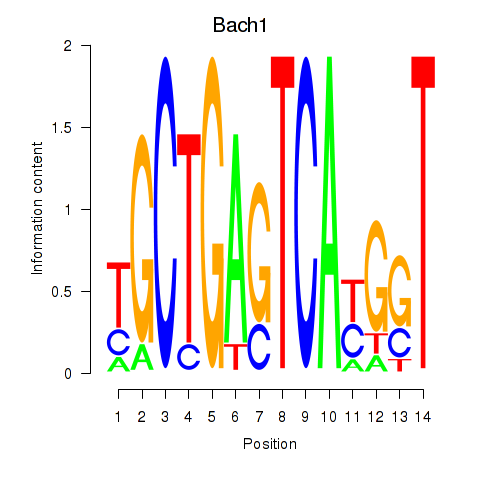
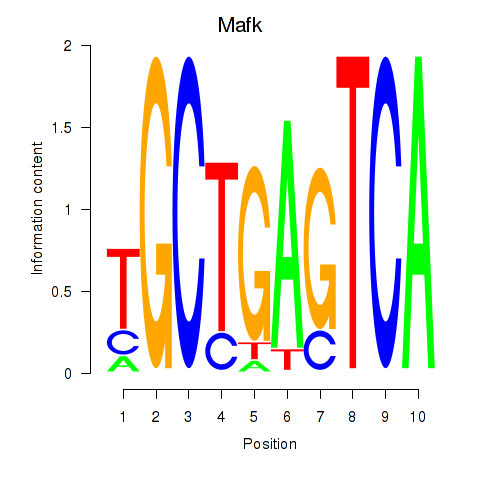
Results for Nfe2_Bach1_Mafk
Z-value: 1.38
Transcription factors associated with Nfe2_Bach1_Mafk
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfe2
|
ENSMUSG00000058794.6 | nuclear factor, erythroid derived 2 |
|
Bach1
|
ENSMUSG00000025612.5 | BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
|
Mafk
|
ENSMUSG00000018143.4 | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein K (avian) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfe2 | mm10_v2_chr15_-_103252810_103252829 | -0.82 | 9.8e-10 | Click! |
| Mafk | mm10_v2_chr5_+_139791513_139791539 | -0.76 | 7.2e-08 | Click! |
| Bach1 | mm10_v2_chr16_+_87698904_87698959 | -0.41 | 1.4e-02 | Click! |
Activity profile of Nfe2_Bach1_Mafk motif
Sorted Z-values of Nfe2_Bach1_Mafk motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_86695897 | 18.29 |
ENSMUST00000034989.8
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr1_+_58029931 | 12.80 |
ENSMUST00000001027.6
|
Aox1
|
aldehyde oxidase 1 |
| chr19_+_12633507 | 12.37 |
ENSMUST00000119960.1
|
Glyat
|
glycine-N-acyltransferase |
| chr9_+_86695542 | 10.68 |
ENSMUST00000150367.2
|
A330041J22Rik
|
RIKEN cDNA A330041J22 gene |
| chr5_-_108675569 | 8.14 |
ENSMUST00000051757.7
|
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1 |
| chr19_+_12633303 | 8.12 |
ENSMUST00000044976.5
|
Glyat
|
glycine-N-acyltransferase |
| chr1_+_165769392 | 7.15 |
ENSMUST00000040298.4
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr7_+_127800604 | 6.46 |
ENSMUST00000046863.5
ENSMUST00000106272.1 ENSMUST00000139068.1 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr5_-_24758008 | 6.44 |
ENSMUST00000047119.4
|
Crygn
|
crystallin, gamma N |
| chr14_+_66635251 | 6.31 |
ENSMUST00000159365.1
ENSMUST00000054661.1 ENSMUST00000159068.1 |
Adra1a
|
adrenergic receptor, alpha 1a |
| chr3_+_123267445 | 5.29 |
ENSMUST00000047923.7
|
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr8_+_119437118 | 5.22 |
ENSMUST00000152420.1
ENSMUST00000098365.3 |
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr8_+_105269837 | 5.04 |
ENSMUST00000172525.1
ENSMUST00000174837.1 ENSMUST00000173859.1 |
Hsf4
|
heat shock transcription factor 4 |
| chr11_+_80428598 | 5.00 |
ENSMUST00000173938.1
ENSMUST00000017572.7 |
Psmd11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr8_+_105269788 | 4.92 |
ENSMUST00000036127.2
ENSMUST00000163734.2 |
Hsf4
|
heat shock transcription factor 4 |
| chr16_+_90220742 | 4.76 |
ENSMUST00000023707.9
|
Sod1
|
superoxide dismutase 1, soluble |
| chr6_-_72235559 | 4.73 |
ENSMUST00000042646.7
|
Atoh8
|
atonal homolog 8 (Drosophila) |
| chrX_+_164269371 | 4.71 |
ENSMUST00000145412.1
ENSMUST00000033749.7 |
Pir
|
pirin |
| chr19_-_4037910 | 4.61 |
ENSMUST00000169613.1
|
Gstp1
|
glutathione S-transferase, pi 1 |
| chr15_-_82407187 | 4.59 |
ENSMUST00000072776.3
|
Cyp2d10
|
cytochrome P450, family 2, subfamily d, polypeptide 10 |
| chr4_+_116685859 | 4.53 |
ENSMUST00000129315.1
ENSMUST00000106470.1 |
Prdx1
|
peroxiredoxin 1 |
| chr4_+_116685544 | 4.51 |
ENSMUST00000135573.1
ENSMUST00000151129.1 |
Prdx1
|
peroxiredoxin 1 |
| chr11_-_50210765 | 4.49 |
ENSMUST00000143379.1
ENSMUST00000015981.5 ENSMUST00000102774.4 |
Sqstm1
|
sequestosome 1 |
| chr19_-_4042165 | 4.41 |
ENSMUST00000042700.9
|
Gstp2
|
glutathione S-transferase, pi 2 |
| chr8_-_93229517 | 4.37 |
ENSMUST00000176282.1
ENSMUST00000034173.7 |
Ces1e
|
carboxylesterase 1E |
| chr18_+_24653691 | 4.37 |
ENSMUST00000068006.7
|
Mocos
|
molybdenum cofactor sulfurase |
| chr2_-_48457245 | 4.35 |
ENSMUST00000127672.1
|
Gm13481
|
predicted gene 13481 |
| chr7_+_127800844 | 4.30 |
ENSMUST00000106271.1
ENSMUST00000138432.1 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr1_+_33908172 | 4.13 |
ENSMUST00000182513.1
ENSMUST00000183034.1 |
Dst
|
dystonin |
| chr19_+_8839298 | 4.10 |
ENSMUST00000160556.1
|
Bscl2
|
Bernardinelli-Seip congenital lipodystrophy 2 homolog (human) |
| chr5_-_65428354 | 4.07 |
ENSMUST00000131263.1
|
Ugdh
|
UDP-glucose dehydrogenase |
| chr10_+_128933782 | 3.88 |
ENSMUST00000099112.2
|
Itga7
|
integrin alpha 7 |
| chr2_+_25428699 | 3.80 |
ENSMUST00000102919.3
|
Abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr11_-_69805617 | 3.78 |
ENSMUST00000051025.4
|
Tmem102
|
transmembrane protein 102 |
| chr11_-_120630516 | 3.71 |
ENSMUST00000106181.1
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr1_-_180199587 | 3.67 |
ENSMUST00000161743.1
|
Adck3
|
aarF domain containing kinase 3 |
| chr16_+_20548577 | 3.67 |
ENSMUST00000003319.5
|
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr13_-_54611274 | 3.56 |
ENSMUST00000049575.7
|
Cltb
|
clathrin, light polypeptide (Lcb) |
| chr10_+_60277627 | 3.51 |
ENSMUST00000105465.1
ENSMUST00000177779.1 ENSMUST00000179238.1 ENSMUST00000004316.8 |
Psap
|
prosaposin |
| chr1_+_107589997 | 3.45 |
ENSMUST00000112706.2
ENSMUST00000000514.4 |
Serpinb8
|
serine (or cysteine) peptidase inhibitor, clade B, member 8 |
| chr16_-_22811399 | 3.30 |
ENSMUST00000040592.4
|
Crygs
|
crystallin, gamma S |
| chr13_-_54611332 | 3.27 |
ENSMUST00000091609.4
|
Cltb
|
clathrin, light polypeptide (Lcb) |
| chr14_+_29018205 | 3.24 |
ENSMUST00000055662.2
|
Lrtm1
|
leucine-rich repeats and transmembrane domains 1 |
| chr2_+_155775333 | 3.16 |
ENSMUST00000029141.5
|
Mmp24
|
matrix metallopeptidase 24 |
| chrX_+_98149666 | 3.16 |
ENSMUST00000052837.7
|
Ar
|
androgen receptor |
| chr12_-_84400929 | 3.08 |
ENSMUST00000122194.1
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr4_-_42168603 | 3.07 |
ENSMUST00000098121.3
|
Gm13305
|
predicted gene 13305 |
| chr12_-_84400851 | 3.04 |
ENSMUST00000117286.1
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr13_-_58215615 | 2.97 |
ENSMUST00000058735.5
ENSMUST00000076454.6 |
Ubqln1
|
ubiquilin 1 |
| chr2_+_122765237 | 2.87 |
ENSMUST00000005953.4
|
Sqrdl
|
sulfide quinone reductase-like (yeast) |
| chr11_-_50325599 | 2.87 |
ENSMUST00000179865.1
ENSMUST00000020637.8 |
Canx
|
calnexin |
| chr4_-_19922599 | 2.86 |
ENSMUST00000029900.5
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr15_+_10223974 | 2.84 |
ENSMUST00000128450.1
ENSMUST00000148257.1 ENSMUST00000128921.1 |
Prlr
|
prolactin receptor |
| chr2_+_11705712 | 2.83 |
ENSMUST00000138856.1
ENSMUST00000078834.5 ENSMUST00000114834.3 ENSMUST00000114833.3 ENSMUST00000114831.2 ENSMUST00000114832.2 |
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr15_-_98677451 | 2.81 |
ENSMUST00000120997.1
ENSMUST00000109149.2 ENSMUST00000003451.4 |
Rnd1
|
Rho family GTPase 1 |
| chr11_+_121146143 | 2.72 |
ENSMUST00000039088.8
ENSMUST00000155694.1 |
Tex19.1
|
testis expressed gene 19.1 |
| chr7_+_143475094 | 2.64 |
ENSMUST00000105917.2
|
Slc22a18
|
solute carrier family 22 (organic cation transporter), member 18 |
| chr2_+_11705355 | 2.64 |
ENSMUST00000128156.2
|
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr2_+_11705437 | 2.57 |
ENSMUST00000148748.1
|
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr19_+_26753588 | 2.55 |
ENSMUST00000177116.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_-_86518578 | 2.54 |
ENSMUST00000134179.1
|
Tmprss11g
|
transmembrane protease, serine 11g |
| chr1_-_173367638 | 2.47 |
ENSMUST00000005470.4
ENSMUST00000111220.1 |
Cadm3
|
cell adhesion molecule 3 |
| chr18_+_61639542 | 2.46 |
ENSMUST00000183083.1
ENSMUST00000183087.1 |
Gm20748
|
predicted gene, 20748 |
| chr3_+_81999461 | 2.45 |
ENSMUST00000107736.1
|
Asic5
|
acid-sensing (proton-gated) ion channel family member 5 |
| chr2_+_11705459 | 2.44 |
ENSMUST00000126394.1
|
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr6_+_138141569 | 2.38 |
ENSMUST00000118091.1
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr13_-_12461432 | 2.35 |
ENSMUST00000143693.1
ENSMUST00000144283.1 ENSMUST00000099820.3 ENSMUST00000135166.1 |
Lgals8
|
lectin, galactose binding, soluble 8 |
| chr5_+_30814571 | 2.33 |
ENSMUST00000031058.8
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr9_+_7272514 | 2.30 |
ENSMUST00000015394.8
|
Mmp13
|
matrix metallopeptidase 13 |
| chr9_-_106476372 | 2.30 |
ENSMUST00000123555.1
ENSMUST00000125850.1 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr11_+_120673359 | 2.29 |
ENSMUST00000135346.1
ENSMUST00000127269.1 ENSMUST00000131727.2 ENSMUST00000149389.1 ENSMUST00000153346.1 |
Aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
| chr5_+_30814722 | 2.28 |
ENSMUST00000114724.1
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr2_+_152105722 | 2.24 |
ENSMUST00000099225.2
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr6_+_17463927 | 2.23 |
ENSMUST00000115442.1
|
Met
|
met proto-oncogene |
| chr2_-_102400863 | 2.19 |
ENSMUST00000102573.1
|
Trim44
|
tripartite motif-containing 44 |
| chr3_+_138217814 | 2.16 |
ENSMUST00000090171.5
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr4_-_57956283 | 2.15 |
ENSMUST00000030051.5
|
Txn1
|
thioredoxin 1 |
| chr2_-_163419508 | 2.11 |
ENSMUST00000046908.3
|
Oser1
|
oxidative stress responsive serine rich 1 |
| chr7_+_127244511 | 2.07 |
ENSMUST00000052509.4
|
Zfp771
|
zinc finger protein 771 |
| chr7_-_44670820 | 2.06 |
ENSMUST00000048102.7
|
Myh14
|
myosin, heavy polypeptide 14 |
| chr2_+_11705287 | 2.01 |
ENSMUST00000135341.1
ENSMUST00000138349.1 ENSMUST00000123600.2 |
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr10_+_18845071 | 1.98 |
ENSMUST00000019998.7
|
Perp
|
PERP, TP53 apoptosis effector |
| chr2_+_109917639 | 1.97 |
ENSMUST00000046548.7
ENSMUST00000111037.2 |
Lgr4
|
leucine-rich repeat-containing G protein-coupled receptor 4 |
| chrX_-_136868537 | 1.97 |
ENSMUST00000058814.6
|
Rab9b
|
RAB9B, member RAS oncogene family |
| chr9_-_106476590 | 1.94 |
ENSMUST00000112479.2
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr2_+_160880642 | 1.94 |
ENSMUST00000109456.2
|
Lpin3
|
lipin 3 |
| chr3_-_89393629 | 1.93 |
ENSMUST00000124783.1
ENSMUST00000126027.1 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr15_-_76014318 | 1.93 |
ENSMUST00000060807.5
|
Fam83h
|
family with sequence similarity 83, member H |
| chr11_-_30649510 | 1.91 |
ENSMUST00000074613.3
|
Acyp2
|
acylphosphatase 2, muscle type |
| chr15_-_74728011 | 1.91 |
ENSMUST00000023261.2
|
Slurp1
|
secreted Ly6/Plaur domain containing 1 |
| chr12_+_70974621 | 1.87 |
ENSMUST00000160027.1
ENSMUST00000160864.1 |
Psma3
|
proteasome (prosome, macropain) subunit, alpha type 3 |
| chr11_+_120530688 | 1.86 |
ENSMUST00000026119.7
|
Gcgr
|
glucagon receptor |
| chr11_-_120630126 | 1.84 |
ENSMUST00000106180.1
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr18_-_34954302 | 1.83 |
ENSMUST00000025217.8
|
Hspa9
|
heat shock protein 9 |
| chr8_-_3717547 | 1.83 |
ENSMUST00000058040.6
|
Gm9814
|
predicted gene 9814 |
| chr6_+_17463749 | 1.82 |
ENSMUST00000115443.1
|
Met
|
met proto-oncogene |
| chr9_-_43239816 | 1.77 |
ENSMUST00000034512.5
|
Oaf
|
OAF homolog (Drosophila) |
| chr5_+_34525797 | 1.76 |
ENSMUST00000125817.1
ENSMUST00000067638.7 |
Sh3bp2
|
SH3-domain binding protein 2 |
| chrX_+_74424632 | 1.75 |
ENSMUST00000114129.2
ENSMUST00000132749.1 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr11_+_60728398 | 1.74 |
ENSMUST00000018743.4
|
Mief2
|
mitochondrial elongation factor 2 |
| chrX_+_74424534 | 1.73 |
ENSMUST00000135165.1
ENSMUST00000114128.1 ENSMUST00000114133.2 ENSMUST00000004330.3 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr9_-_21239310 | 1.72 |
ENSMUST00000164812.1
ENSMUST00000049567.4 |
Keap1
|
kelch-like ECH-associated protein 1 |
| chr15_-_76195710 | 1.70 |
ENSMUST00000023226.6
|
Plec
|
plectin |
| chrX_+_133908441 | 1.70 |
ENSMUST00000113304.1
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr17_-_24644933 | 1.68 |
ENSMUST00000019684.5
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr12_-_85288419 | 1.68 |
ENSMUST00000121930.1
|
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr1_+_191978247 | 1.68 |
ENSMUST00000175680.1
|
Rd3
|
retinal degeneration 3 |
| chr10_+_61648552 | 1.66 |
ENSMUST00000020286.6
|
Ppa1
|
pyrophosphatase (inorganic) 1 |
| chr11_+_78178105 | 1.65 |
ENSMUST00000147819.1
|
Tlcd1
|
TLC domain containing 1 |
| chr7_+_101321703 | 1.65 |
ENSMUST00000174291.1
ENSMUST00000167888.2 ENSMUST00000172662.1 ENSMUST00000173270.1 ENSMUST00000174083.1 |
Stard10
|
START domain containing 10 |
| chr15_-_100599983 | 1.64 |
ENSMUST00000073837.6
|
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr10_-_81427114 | 1.62 |
ENSMUST00000078185.7
ENSMUST00000020461.8 ENSMUST00000105321.3 |
Nfic
|
nuclear factor I/C |
| chr9_-_107679592 | 1.61 |
ENSMUST00000010205.7
|
Gnat1
|
guanine nucleotide binding protein, alpha transducing 1 |
| chr5_+_102845007 | 1.60 |
ENSMUST00000070000.4
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr3_-_84220853 | 1.60 |
ENSMUST00000154152.1
ENSMUST00000107693.2 ENSMUST00000107695.2 |
Trim2
|
tripartite motif-containing 2 |
| chr9_+_32224457 | 1.60 |
ENSMUST00000183121.1
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr5_+_147860615 | 1.59 |
ENSMUST00000031654.6
|
Pomp
|
proteasome maturation protein |
| chr9_-_106476104 | 1.59 |
ENSMUST00000156426.1
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr6_+_124996681 | 1.53 |
ENSMUST00000032479.4
|
Pianp
|
PILR alpha associated neural protein |
| chr5_+_43233928 | 1.53 |
ENSMUST00000114066.1
ENSMUST00000114065.1 |
Cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr2_+_163547148 | 1.52 |
ENSMUST00000109411.1
ENSMUST00000018094.6 |
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr11_-_116189542 | 1.52 |
ENSMUST00000148601.1
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr7_-_104315455 | 1.52 |
ENSMUST00000106837.1
ENSMUST00000106839.2 ENSMUST00000070943.6 |
Trim12a
|
tripartite motif-containing 12A |
| chr9_+_65214690 | 1.51 |
ENSMUST00000069000.7
|
Parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr3_-_89393294 | 1.51 |
ENSMUST00000142119.1
ENSMUST00000029677.8 ENSMUST00000148361.1 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr17_-_24220738 | 1.50 |
ENSMUST00000024930.7
|
1600002H07Rik
|
RIKEN cDNA 1600002H07 gene |
| chr10_+_4611971 | 1.49 |
ENSMUST00000105590.1
ENSMUST00000067086.7 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr2_-_156004147 | 1.48 |
ENSMUST00000156993.1
ENSMUST00000141437.1 |
6430550D23Rik
|
RIKEN cDNA 6430550D23 gene |
| chr19_+_57611020 | 1.48 |
ENSMUST00000077282.5
|
Atrnl1
|
attractin like 1 |
| chr6_+_17463826 | 1.47 |
ENSMUST00000140070.1
|
Met
|
met proto-oncogene |
| chr19_+_38481057 | 1.46 |
ENSMUST00000182481.1
|
Plce1
|
phospholipase C, epsilon 1 |
| chr6_-_83656082 | 1.44 |
ENSMUST00000014686.2
|
Clec4f
|
C-type lectin domain family 4, member f |
| chr4_+_150855064 | 1.43 |
ENSMUST00000030811.1
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr2_+_91257323 | 1.42 |
ENSMUST00000111349.2
ENSMUST00000131711.1 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr11_-_98438941 | 1.42 |
ENSMUST00000002655.7
|
Mien1
|
migration and invasion enhancer 1 |
| chr10_+_128058974 | 1.41 |
ENSMUST00000084771.2
|
Ptges3
|
prostaglandin E synthase 3 (cytosolic) |
| chr13_-_23710714 | 1.39 |
ENSMUST00000091707.6
ENSMUST00000006787.7 ENSMUST00000091706.6 |
Hfe
|
hemochromatosis |
| chr9_+_32224246 | 1.38 |
ENSMUST00000168954.2
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr5_+_43233463 | 1.37 |
ENSMUST00000169035.1
ENSMUST00000166713.1 |
Cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr2_-_160313616 | 1.37 |
ENSMUST00000109475.2
|
Gm826
|
predicted gene 826 |
| chr10_+_128058947 | 1.34 |
ENSMUST00000052798.7
|
Ptges3
|
prostaglandin E synthase 3 (cytosolic) |
| chr9_-_121916288 | 1.30 |
ENSMUST00000062474.4
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr12_-_108275409 | 1.30 |
ENSMUST00000136175.1
|
Ccdc85c
|
coiled-coil domain containing 85C |
| chr15_-_76206309 | 1.29 |
ENSMUST00000073418.6
ENSMUST00000171634.1 ENSMUST00000076442.5 |
Plec
|
plectin |
| chr3_-_116968827 | 1.29 |
ENSMUST00000119557.1
|
Palmd
|
palmdelphin |
| chr18_-_63692341 | 1.28 |
ENSMUST00000025476.2
|
Txnl1
|
thioredoxin-like 1 |
| chr15_-_100599864 | 1.27 |
ENSMUST00000177247.2
ENSMUST00000177505.2 |
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr1_-_134235420 | 1.27 |
ENSMUST00000038191.6
ENSMUST00000086465.4 |
Adora1
|
adenosine A1 receptor |
| chr7_-_18616498 | 1.25 |
ENSMUST00000057810.6
|
Psg23
|
pregnancy-specific glycoprotein 23 |
| chr10_-_25536114 | 1.24 |
ENSMUST00000179685.1
|
Smlr1
|
small leucine-rich protein 1 |
| chr8_+_94532990 | 1.19 |
ENSMUST00000048653.2
ENSMUST00000109537.1 |
Cpne2
|
copine II |
| chr14_-_18239053 | 1.17 |
ENSMUST00000090543.5
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr7_+_49759100 | 1.17 |
ENSMUST00000085272.5
|
Htatip2
|
HIV-1 tat interactive protein 2, homolog (human) |
| chr5_-_49524809 | 1.17 |
ENSMUST00000087395.4
|
Kcnip4
|
Kv channel interacting protein 4 |
| chr2_+_136713444 | 1.15 |
ENSMUST00000028727.4
ENSMUST00000110098.3 |
Snap25
|
synaptosomal-associated protein 25 |
| chr15_+_80255184 | 1.14 |
ENSMUST00000109605.3
|
Atf4
|
activating transcription factor 4 |
| chr10_-_34418465 | 1.14 |
ENSMUST00000099973.3
ENSMUST00000105512.1 ENSMUST00000047885.7 |
Nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr10_+_43901782 | 1.13 |
ENSMUST00000054418.5
|
Rtn4ip1
|
reticulon 4 interacting protein 1 |
| chrX_+_133908418 | 1.13 |
ENSMUST00000033606.8
ENSMUST00000113303.1 ENSMUST00000165805.1 |
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr8_+_105326354 | 1.12 |
ENSMUST00000015000.5
ENSMUST00000098453.2 |
Tmem208
|
transmembrane protein 208 |
| chr12_-_80436571 | 1.11 |
ENSMUST00000054145.6
|
Dcaf5
|
DDB1 and CUL4 associated factor 5 |
| chr1_-_120074023 | 1.11 |
ENSMUST00000056089.7
|
Tmem37
|
transmembrane protein 37 |
| chr11_+_120673018 | 1.11 |
ENSMUST00000106158.2
ENSMUST00000103016.1 ENSMUST00000168714.1 |
Aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
| chr19_+_5877794 | 1.09 |
ENSMUST00000145200.1
ENSMUST00000025732.7 ENSMUST00000125114.1 ENSMUST00000155697.1 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr13_-_85127514 | 1.06 |
ENSMUST00000179230.1
|
Gm4076
|
predicted gene 4076 |
| chr5_+_22550391 | 1.06 |
ENSMUST00000181374.1
ENSMUST00000181764.1 ENSMUST00000181209.1 |
6030443J06Rik
|
RIKEN cDNA 6030443J06 gene |
| chr15_+_3270767 | 1.06 |
ENSMUST00000082424.4
ENSMUST00000159158.1 ENSMUST00000159216.1 ENSMUST00000160311.1 |
Sepp1
|
selenoprotein P, plasma, 1 |
| chrX_-_97377190 | 1.06 |
ENSMUST00000037353.3
|
Eda2r
|
ectodysplasin A2 receptor |
| chr18_+_84088077 | 1.06 |
ENSMUST00000060223.2
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr2_+_30266721 | 1.04 |
ENSMUST00000113645.1
ENSMUST00000133877.1 ENSMUST00000139719.1 ENSMUST00000113643.1 ENSMUST00000150695.1 |
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr14_-_20794009 | 1.03 |
ENSMUST00000100837.3
ENSMUST00000080440.6 ENSMUST00000071816.6 |
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr14_-_37135126 | 1.02 |
ENSMUST00000042564.9
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr2_+_180171485 | 1.02 |
ENSMUST00000061437.4
|
Adrm1
|
adhesion regulating molecule 1 |
| chr9_-_25151772 | 1.02 |
ENSMUST00000008573.7
|
Herpud2
|
HERPUD family member 2 |
| chr2_+_30266513 | 1.02 |
ENSMUST00000091132.6
|
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr19_+_26623419 | 1.00 |
ENSMUST00000176584.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr11_-_48871344 | 1.00 |
ENSMUST00000049519.3
|
Irgm1
|
immunity-related GTPase family M member 1 |
| chr19_+_56287943 | 0.99 |
ENSMUST00000166049.1
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr19_+_56287911 | 0.98 |
ENSMUST00000095948.4
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr11_-_120437656 | 0.98 |
ENSMUST00000044271.8
ENSMUST00000103017.3 |
Nploc4
|
nuclear protein localization 4 homolog (S. cerevisiae) |
| chr9_-_122310921 | 0.97 |
ENSMUST00000180685.1
|
Gm26797
|
predicted gene, 26797 |
| chr7_+_26435614 | 0.97 |
ENSMUST00000119386.1
|
Nlrp4a
|
NLR family, pyrin domain containing 4A |
| chr17_+_26933070 | 0.96 |
ENSMUST00000073724.5
|
Phf1
|
PHD finger protein 1 |
| chr3_-_88552859 | 0.94 |
ENSMUST00000119002.1
ENSMUST00000029698.8 |
Lamtor2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr17_-_45592485 | 0.94 |
ENSMUST00000166119.1
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr12_+_35047180 | 0.94 |
ENSMUST00000048519.9
ENSMUST00000163677.1 |
Snx13
|
sorting nexin 13 |
| chr19_+_42052228 | 0.94 |
ENSMUST00000164518.2
|
4933411K16Rik
|
RIKEN cDNA 4933411K16 gene |
| chr5_-_107875035 | 0.91 |
ENSMUST00000138111.1
ENSMUST00000112642.1 |
Evi5
|
ecotropic viral integration site 5 |
| chr8_-_70506710 | 0.91 |
ENSMUST00000136913.1
ENSMUST00000075175.5 |
2810428I15Rik
|
RIKEN cDNA 2810428I15 gene |
| chr10_+_128790903 | 0.90 |
ENSMUST00000026411.6
|
Mmp19
|
matrix metallopeptidase 19 |
| chr2_+_112261926 | 0.90 |
ENSMUST00000028553.3
|
Nop10
|
NOP10 ribonucleoprotein |
| chr17_-_45592262 | 0.88 |
ENSMUST00000164769.1
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chrX_+_21714896 | 0.88 |
ENSMUST00000033414.7
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr16_+_20651652 | 0.87 |
ENSMUST00000007212.8
|
Psmd2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr19_-_21652779 | 0.87 |
ENSMUST00000179768.1
ENSMUST00000178523.1 ENSMUST00000038830.3 |
1110059E24Rik
|
RIKEN cDNA 1110059E24 gene |
| chr16_-_36990449 | 0.86 |
ENSMUST00000075869.6
|
Fbxo40
|
F-box protein 40 |
| chr11_-_48871408 | 0.86 |
ENSMUST00000097271.2
|
Irgm1
|
immunity-related GTPase family M member 1 |
| chr14_-_21848924 | 0.86 |
ENSMUST00000124549.1
|
Comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr3_+_122245557 | 0.86 |
ENSMUST00000029769.7
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr2_+_180042496 | 0.84 |
ENSMUST00000041126.8
|
Ss18l1
|
synovial sarcoma translocation gene on chromosome 18-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfe2_Bach1_Mafk
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 18.3 | GO:0006108 | malate metabolic process(GO:0006108) regulation of NADP metabolic process(GO:1902031) |
| 2.6 | 12.8 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 2.1 | 6.3 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 1.9 | 5.8 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 1.5 | 4.6 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 1.2 | 10.8 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 1.2 | 4.6 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 1.1 | 3.2 | GO:0060748 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 1.0 | 8.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 1.0 | 3.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.9 | 2.7 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.9 | 3.4 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.8 | 5.5 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.7 | 4.4 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.6 | 2.4 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.6 | 2.3 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.6 | 1.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.5 | 3.8 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.5 | 2.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.5 | 4.8 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.5 | 12.5 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.5 | 8.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.5 | 1.5 | GO:1902569 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.5 | 2.8 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.5 | 1.8 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.4 | 1.3 | GO:0060995 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.4 | 10.0 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.4 | 1.3 | GO:0070256 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.4 | 4.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.4 | 3.6 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.4 | 2.4 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.4 | 1.2 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.4 | 1.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.4 | 1.5 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.4 | 5.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.4 | 2.9 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.4 | 9.0 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.3 | 3.8 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.3 | 1.6 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) |
| 0.3 | 1.9 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.3 | 1.9 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.3 | 1.5 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.3 | 3.5 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.3 | 1.4 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.3 | 1.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.3 | 1.4 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 0.3 | 0.8 | GO:0015881 | creatine transport(GO:0015881) |
| 0.3 | 1.8 | GO:0015862 | uridine transport(GO:0015862) |
| 0.3 | 0.8 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.3 | 1.5 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 0.7 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.2 | 0.7 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.2 | 2.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.2 | 3.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 4.7 | GO:0051450 | myoblast proliferation(GO:0051450) |
| 0.2 | 1.6 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.2 | 1.6 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.2 | 6.1 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) |
| 0.2 | 4.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 1.0 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 2.8 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 0.9 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.2 | 1.6 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 3.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 3.4 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.2 | 4.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 3.4 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.2 | 2.8 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 1.0 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.2 | 1.9 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.4 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 1.2 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 2.0 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 4.7 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.4 | GO:0010819 | regulation of T cell chemotaxis(GO:0010819) |
| 0.1 | 1.4 | GO:0051132 | NK T cell activation(GO:0051132) |
| 0.1 | 0.6 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 1.7 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 1.7 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 0.3 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 0.2 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.3 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 4.1 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.1 | 0.5 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.1 | 4.7 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.8 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 | 0.3 | GO:1903465 | orbitofrontal cortex development(GO:0021769) motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development(GO:0035607) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.1 | 15.8 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 0.7 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.1 | 0.3 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.4 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 1.0 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 2.9 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 1.9 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.1 | 0.3 | GO:0035938 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 0.3 | GO:0060463 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.1 | 1.8 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 0.2 | GO:0035483 | gastric emptying(GO:0035483) |
| 0.1 | 0.5 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 4.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.4 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.3 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 2.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.6 | GO:0002551 | mast cell chemotaxis(GO:0002551) mast cell migration(GO:0097531) |
| 0.1 | 5.7 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 0.5 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 1.7 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 0.3 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 2.6 | GO:0015893 | drug transport(GO:0015893) |
| 0.1 | 1.2 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.1 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 1.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.5 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.1 | 0.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 | 2.2 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.2 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.3 | GO:0035247 | peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.1 | 1.4 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.5 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 3.6 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 1.9 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 2.1 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.0 | 0.5 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 2.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.7 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.8 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.1 | GO:0021995 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 0.4 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 3.3 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.2 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 1.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 2.7 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.2 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.7 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.3 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.2 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:0071321 | cellular response to cGMP(GO:0071321) membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.4 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.6 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.0 | 0.4 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.6 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 1.5 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 2.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.4 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 1.5 | GO:0032835 | glomerulus development(GO:0032835) |
| 0.0 | 1.3 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 1.1 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 1.6 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 1.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.4 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0001712 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.3 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.5 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.8 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 1.9 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 1.1 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.3 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.6 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:2001179 | regulation of interleukin-10 secretion(GO:2001179) |
| 0.0 | 1.0 | GO:0006986 | response to unfolded protein(GO:0006986) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 1.5 | 4.5 | GO:0044753 | amphisome(GO:0044753) |
| 0.7 | 7.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.7 | 4.1 | GO:0031673 | H zone(GO:0031673) |
| 0.6 | 6.8 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.6 | 3.9 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.5 | 1.6 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.4 | 5.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.3 | 2.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.3 | 9.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 3.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.3 | 4.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 3.3 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.3 | 1.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.3 | 1.1 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.2 | 2.5 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 3.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 0.9 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.2 | 2.9 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 1.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 0.6 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.2 | 3.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 3.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 1.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 7.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 3.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 0.9 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 1.9 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 4.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.4 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 6.2 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 0.5 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 1.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 2.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.8 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 2.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 3.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.3 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 1.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.4 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 1.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 1.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.6 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.1 | 0.7 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 3.9 | GO:0044438 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 4.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.9 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 4.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.6 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 1.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 4.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 8.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 2.1 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 1.0 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.0 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 25.9 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 7.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.3 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 9.1 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 1.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.3 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 5.1 | 20.5 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 3.0 | 15.0 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 1.6 | 6.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 1.4 | 5.5 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 1.4 | 4.1 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 1.2 | 6.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 1.2 | 3.5 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 1.0 | 4.8 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.9 | 9.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.9 | 3.6 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.9 | 4.4 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.8 | 10.8 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.7 | 2.7 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.6 | 5.8 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.6 | 1.9 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.6 | 3.5 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.6 | 6.8 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.6 | 2.8 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.5 | 1.6 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.5 | 1.5 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.5 | 1.5 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.5 | 3.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.5 | 8.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.5 | 1.9 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.4 | 12.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.4 | 1.7 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.4 | 1.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.4 | 1.5 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.4 | 7.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.4 | 1.1 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.3 | 1.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.3 | 2.5 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.3 | 0.8 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.3 | 6.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.3 | 1.5 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.2 | 4.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.2 | 1.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 3.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 2.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 1.4 | GO:0005534 | galactose binding(GO:0005534) |
| 0.2 | 0.9 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.2 | 4.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 3.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 2.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 0.8 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.2 | 0.6 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 4.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.3 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 1.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 1.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.9 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 2.0 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.4 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 2.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 1.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 1.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 4.0 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 2.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.6 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 1.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 2.6 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.4 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 3.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.1 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 4.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 3.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 0.2 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.1 | 0.5 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 1.5 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 2.1 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.3 | GO:0034648 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.5 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.5 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 1.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 2.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 2.2 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 13.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 5.3 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 1.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 2.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 3.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.8 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 1.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 9.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 2.8 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 3.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 7.6 | GO:0048037 | cofactor binding(GO:0048037) |
| 0.0 | 3.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 2.7 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.9 | GO:0051213 | dioxygenase activity(GO:0051213) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 11.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 8.0 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 5.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 5.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 8.6 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 2.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 5.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.8 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 1.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 2.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 0.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 3.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.8 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 1.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 12.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 3.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.4 | 8.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.4 | 33.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.4 | 2.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 4.5 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.3 | 14.2 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.2 | 4.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 9.6 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.2 | 6.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 3.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 2.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 19.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 2.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 3.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 1.6 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 7.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 3.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 4.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 8.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 1.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 3.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.1 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 1.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |