Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
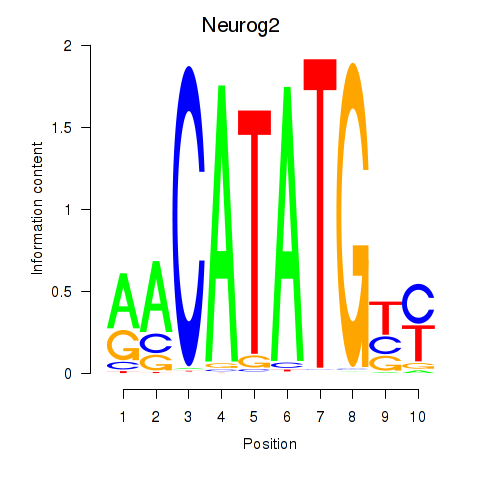
Results for Neurog2
Z-value: 1.17
Transcription factors associated with Neurog2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Neurog2
|
ENSMUSG00000027967.7 | neurogenin 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Neurog2 | mm10_v2_chr3_+_127633134_127633140 | 0.54 | 6.7e-04 | Click! |
Activity profile of Neurog2 motif
Sorted Z-values of Neurog2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_90490714 | 18.35 |
ENSMUST00000042755.3
|
Afp
|
alpha fetoprotein |
| chr11_-_102365111 | 14.50 |
ENSMUST00000006749.9
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr13_+_54701457 | 8.96 |
ENSMUST00000037145.7
|
Cdhr2
|
cadherin-related family member 2 |
| chr4_-_118457450 | 8.23 |
ENSMUST00000106375.1
ENSMUST00000006556.3 ENSMUST00000168404.1 |
Mpl
|
myeloproliferative leukemia virus oncogene |
| chr9_-_70421533 | 8.21 |
ENSMUST00000034742.6
|
Ccnb2
|
cyclin B2 |
| chr4_-_118457509 | 7.83 |
ENSMUST00000102671.3
|
Mpl
|
myeloproliferative leukemia virus oncogene |
| chr11_-_69602741 | 7.45 |
ENSMUST00000138694.1
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr8_-_107065632 | 7.31 |
ENSMUST00000034393.5
|
Tmed6
|
transmembrane emp24 protein transport domain containing 6 |
| chr7_+_30699783 | 6.40 |
ENSMUST00000013227.7
|
2200002J24Rik
|
RIKEN cDNA 2200002J24 gene |
| chr7_-_126704816 | 6.38 |
ENSMUST00000032949.7
|
Coro1a
|
coronin, actin binding protein 1A |
| chrX_+_56454871 | 6.34 |
ENSMUST00000039374.2
ENSMUST00000101553.2 |
Ddx26b
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr10_-_128401218 | 6.21 |
ENSMUST00000042666.5
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr16_-_75909272 | 6.11 |
ENSMUST00000114239.2
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr1_+_170277376 | 5.93 |
ENSMUST00000179976.1
|
Sh2d1b1
|
SH2 domain protein 1B1 |
| chr10_-_128400448 | 5.80 |
ENSMUST00000167859.1
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr11_-_72550255 | 5.77 |
ENSMUST00000021154.6
|
Spns3
|
spinster homolog 3 |
| chr7_-_126704736 | 5.73 |
ENSMUST00000131415.1
|
Coro1a
|
coronin, actin binding protein 1A |
| chr19_+_7268296 | 5.28 |
ENSMUST00000066646.4
|
Rcor2
|
REST corepressor 2 |
| chr11_-_97996171 | 5.20 |
ENSMUST00000042971.9
|
Arl5c
|
ADP-ribosylation factor-like 5C |
| chr7_-_126704522 | 5.16 |
ENSMUST00000135087.1
|
Coro1a
|
coronin, actin binding protein 1A |
| chr6_-_52226165 | 4.69 |
ENSMUST00000114425.2
|
Hoxa9
|
homeobox A9 |
| chr3_-_10301169 | 4.54 |
ENSMUST00000119761.1
ENSMUST00000029043.6 |
Fabp12
|
fatty acid binding protein 12 |
| chr2_+_118772766 | 4.47 |
ENSMUST00000130293.1
ENSMUST00000061360.3 |
Phgr1
|
proline/histidine/glycine-rich 1 |
| chr7_-_127137807 | 4.38 |
ENSMUST00000049931.5
|
Spn
|
sialophorin |
| chr7_-_44748306 | 3.98 |
ENSMUST00000118162.1
ENSMUST00000140599.2 ENSMUST00000120798.1 |
Zfp473
|
zinc finger protein 473 |
| chr15_+_78913916 | 3.54 |
ENSMUST00000089378.4
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr19_+_52264323 | 3.46 |
ENSMUST00000039652.4
|
Ins1
|
insulin I |
| chr2_+_24345282 | 3.41 |
ENSMUST00000114485.2
|
Il1rn
|
interleukin 1 receptor antagonist |
| chr9_+_106281061 | 3.21 |
ENSMUST00000072206.6
|
Poc1a
|
POC1 centriolar protein homolog A (Chlamydomonas) |
| chr6_-_128275577 | 3.19 |
ENSMUST00000130454.1
|
Tead4
|
TEA domain family member 4 |
| chr6_-_83033422 | 3.19 |
ENSMUST00000089651.5
|
Dok1
|
docking protein 1 |
| chr13_+_49544443 | 3.15 |
ENSMUST00000177948.1
ENSMUST00000021820.6 |
Aspn
|
asporin |
| chr19_-_53371766 | 3.11 |
ENSMUST00000086887.1
|
Gm10197
|
predicted gene 10197 |
| chr2_+_24345305 | 3.10 |
ENSMUST00000114482.1
|
Il1rn
|
interleukin 1 receptor antagonist |
| chr4_+_127172866 | 3.10 |
ENSMUST00000106094.2
|
Dlgap3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr3_-_126998408 | 3.07 |
ENSMUST00000182764.1
ENSMUST00000044443.8 |
Ank2
|
ankyrin 2, brain |
| chr9_-_49798905 | 2.96 |
ENSMUST00000114476.2
|
Ncam1
|
neural cell adhesion molecule 1 |
| chr18_+_50051702 | 2.91 |
ENSMUST00000134348.1
ENSMUST00000153873.2 |
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr2_-_105399286 | 2.90 |
ENSMUST00000006128.6
|
Rcn1
|
reticulocalbin 1 |
| chr6_+_41354105 | 2.87 |
ENSMUST00000072103.5
|
Try10
|
trypsin 10 |
| chr13_-_44455303 | 2.86 |
ENSMUST00000183264.1
|
Gm27007
|
predicted gene, 27007 |
| chr11_+_68503019 | 2.77 |
ENSMUST00000102613.1
ENSMUST00000060441.6 |
Pik3r6
|
phosphoinositide-3-kinase, regulatory subunit 6 |
| chr7_+_131032061 | 2.68 |
ENSMUST00000084509.3
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr15_-_91191733 | 2.57 |
ENSMUST00000069511.6
|
Abcd2
|
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr7_-_44748049 | 2.57 |
ENSMUST00000120074.1
|
Zfp473
|
zinc finger protein 473 |
| chr3_+_90526849 | 2.54 |
ENSMUST00000167598.2
ENSMUST00000164481.2 |
S100a14
|
S100 calcium binding protein A14 |
| chr1_-_155232710 | 2.51 |
ENSMUST00000035914.3
|
BC034090
|
cDNA sequence BC034090 |
| chr9_+_86743641 | 2.44 |
ENSMUST00000179574.1
|
Prss35
|
protease, serine, 35 |
| chr10_+_74967164 | 2.30 |
ENSMUST00000037813.4
|
Gnaz
|
guanine nucleotide binding protein, alpha z subunit |
| chr19_-_53589067 | 2.22 |
ENSMUST00000095978.3
|
Nutf2-ps1
|
nuclear transport factor 2, pseudogene 1 |
| chr2_-_163397946 | 2.17 |
ENSMUST00000017961.4
ENSMUST00000109425.2 |
Jph2
|
junctophilin 2 |
| chr17_-_27167759 | 2.14 |
ENSMUST00000025046.2
|
Ip6k3
|
inositol hexaphosphate kinase 3 |
| chr10_+_128232065 | 2.13 |
ENSMUST00000055539.4
ENSMUST00000105244.1 ENSMUST00000105243.2 ENSMUST00000125289.1 ENSMUST00000105242.1 |
Timeless
|
timeless circadian clock 1 |
| chr9_+_108560422 | 2.08 |
ENSMUST00000081111.8
|
Impdh2
|
inosine 5'-phosphate dehydrogenase 2 |
| chr9_+_86743616 | 2.07 |
ENSMUST00000036426.6
|
Prss35
|
protease, serine, 35 |
| chr12_-_78980758 | 2.04 |
ENSMUST00000174072.1
|
Tmem229b
|
transmembrane protein 229B |
| chrX_+_164373363 | 2.04 |
ENSMUST00000033751.7
|
Figf
|
c-fos induced growth factor |
| chr11_+_58954675 | 2.01 |
ENSMUST00000108817.3
ENSMUST00000047697.5 |
Hist3h2a
Trim17
|
histone cluster 3, H2a tripartite motif-containing 17 |
| chr9_-_60649793 | 1.89 |
ENSMUST00000053171.7
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr14_-_70167973 | 1.86 |
ENSMUST00000125300.1
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr13_-_97747399 | 1.78 |
ENSMUST00000144993.1
|
5330416C01Rik
|
RIKEN cDNA 5330416C01 gene |
| chr1_+_180935022 | 1.76 |
ENSMUST00000037361.8
|
Lefty1
|
left right determination factor 1 |
| chr9_-_21918089 | 1.69 |
ENSMUST00000128442.1
ENSMUST00000119055.1 ENSMUST00000122211.1 ENSMUST00000115351.3 |
Rab3d
|
RAB3D, member RAS oncogene family |
| chr7_+_30314810 | 1.69 |
ENSMUST00000054594.8
ENSMUST00000177078.1 ENSMUST00000176504.1 ENSMUST00000176304.1 |
Syne4
|
spectrin repeat containing, nuclear envelope family member 4 |
| chr15_-_76918010 | 1.68 |
ENSMUST00000048854.7
|
Zfp647
|
zinc finger protein 647 |
| chr11_+_69015911 | 1.62 |
ENSMUST00000021278.7
ENSMUST00000161455.1 ENSMUST00000116359.2 |
Ctc1
|
CTS telomere maintenance complex component 1 |
| chr3_-_59220150 | 1.59 |
ENSMUST00000170388.1
|
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr6_-_139501907 | 1.57 |
ENSMUST00000170650.1
|
Rergl
|
RERG/RAS-like |
| chrX_-_111537947 | 1.57 |
ENSMUST00000132319.1
ENSMUST00000123951.1 |
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr6_+_147032528 | 1.56 |
ENSMUST00000036194.4
|
Rep15
|
RAB15 effector protein |
| chr13_-_97747373 | 1.53 |
ENSMUST00000123535.1
|
5330416C01Rik
|
RIKEN cDNA 5330416C01 gene |
| chr2_+_163694015 | 1.53 |
ENSMUST00000109400.2
|
Pkig
|
protein kinase inhibitor, gamma |
| chr2_+_57997884 | 1.51 |
ENSMUST00000112616.1
ENSMUST00000166729.1 |
Galnt5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 |
| chr7_+_28788955 | 1.48 |
ENSMUST00000059857.7
|
Rinl
|
Ras and Rab interactor-like |
| chrX_+_56786527 | 1.47 |
ENSMUST00000144600.1
|
Fhl1
|
four and a half LIM domains 1 |
| chr2_+_75832168 | 1.45 |
ENSMUST00000047232.7
ENSMUST00000111952.2 |
Agps
|
alkylglycerone phosphate synthase |
| chr2_+_150323702 | 1.42 |
ENSMUST00000133235.2
|
Gm10130
|
predicted gene 10130 |
| chr17_-_35074485 | 1.37 |
ENSMUST00000007259.3
|
Ly6g6d
|
lymphocyte antigen 6 complex, locus G6D |
| chr7_+_4925802 | 1.35 |
ENSMUST00000057612.7
|
Ssc5d
|
scavenger receptor cysteine rich domain containing (5 domains) |
| chr9_-_75441652 | 1.33 |
ENSMUST00000181896.1
|
A130057D12Rik
|
RIKEN cDNA A130057D12 gene |
| chr1_+_136017967 | 1.32 |
ENSMUST00000063719.8
ENSMUST00000118832.1 |
Tmem9
|
transmembrane protein 9 |
| chr9_-_58158498 | 1.30 |
ENSMUST00000168864.2
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr10_+_69534039 | 1.30 |
ENSMUST00000182557.1
|
Ank3
|
ankyrin 3, epithelial |
| chr13_-_62607499 | 1.26 |
ENSMUST00000091563.4
|
6720489N17Rik
|
RIKEN cDNA 6720489N17 gene |
| chr8_+_94667082 | 1.25 |
ENSMUST00000109527.4
|
Arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chrX_-_7188713 | 1.25 |
ENSMUST00000004428.7
|
Clcn5
|
chloride channel 5 |
| chr14_+_58893465 | 1.23 |
ENSMUST00000079960.1
|
Rpl13-ps3
|
ribosomal protein L13, pseudogene 3 |
| chr7_-_64392214 | 1.23 |
ENSMUST00000032735.5
|
Mphosph10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr11_+_120232921 | 1.22 |
ENSMUST00000122148.1
ENSMUST00000044985.7 |
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chr9_-_71896047 | 1.22 |
ENSMUST00000184448.1
|
Tcf12
|
transcription factor 12 |
| chr8_+_94666722 | 1.20 |
ENSMUST00000034228.8
|
Arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr16_+_57353093 | 1.20 |
ENSMUST00000159816.1
|
Filip1l
|
filamin A interacting protein 1-like |
| chr10_-_70655934 | 1.20 |
ENSMUST00000162144.1
ENSMUST00000162793.1 |
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like |
| chr3_+_18054258 | 1.19 |
ENSMUST00000026120.6
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr3_+_3508024 | 1.14 |
ENSMUST00000108393.1
ENSMUST00000017832.8 |
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr10_+_4266323 | 1.10 |
ENSMUST00000045730.5
|
Akap12
|
A kinase (PRKA) anchor protein (gravin) 12 |
| chr19_+_37207528 | 1.09 |
ENSMUST00000024078.7
ENSMUST00000112391.1 |
March5
|
membrane-associated ring finger (C3HC4) 5 |
| chr2_-_79456750 | 1.07 |
ENSMUST00000041099.4
|
Neurod1
|
neurogenic differentiation 1 |
| chr1_+_93373874 | 1.06 |
ENSMUST00000058682.4
|
Ano7
|
anoctamin 7 |
| chr13_+_37345338 | 1.05 |
ENSMUST00000021860.5
|
Ly86
|
lymphocyte antigen 86 |
| chr11_+_73160403 | 1.05 |
ENSMUST00000006104.3
|
P2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr17_-_24443093 | 1.04 |
ENSMUST00000088506.5
|
Dnase1l2
|
deoxyribonuclease 1-like 2 |
| chr11_-_103938211 | 1.03 |
ENSMUST00000133774.2
ENSMUST00000149642.1 |
Nsf
|
N-ethylmaleimide sensitive fusion protein |
| chr4_-_82850721 | 1.02 |
ENSMUST00000139401.1
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr2_+_86007778 | 1.01 |
ENSMUST00000062166.1
|
Olfr1032
|
olfactory receptor 1032 |
| chr1_+_150392794 | 1.01 |
ENSMUST00000124973.2
|
Tpr
|
translocated promoter region |
| chr6_+_112273758 | 1.00 |
ENSMUST00000032376.5
|
Lmcd1
|
LIM and cysteine-rich domains 1 |
| chr6_-_122340499 | 0.99 |
ENSMUST00000160843.1
|
Phc1
|
polyhomeotic-like 1 (Drosophila) |
| chr4_-_141598206 | 0.98 |
ENSMUST00000131317.1
ENSMUST00000006381.4 ENSMUST00000129602.1 |
Fblim1
|
filamin binding LIM protein 1 |
| chr16_+_10835046 | 0.98 |
ENSMUST00000037913.8
|
Rmi2
|
RMI2, RecQ mediated genome instability 2, homolog (S. cerevisiae) |
| chr13_+_21180179 | 0.98 |
ENSMUST00000021761.5
|
Trim27
|
tripartite motif-containing 27 |
| chr1_-_111864869 | 0.94 |
ENSMUST00000035462.5
|
Dsel
|
dermatan sulfate epimerase-like |
| chr8_-_122476036 | 0.93 |
ENSMUST00000014614.3
|
Rnf166
|
ring finger protein 166 |
| chr12_+_108410625 | 0.92 |
ENSMUST00000109857.1
|
Eml1
|
echinoderm microtubule associated protein like 1 |
| chrX_+_107148927 | 0.91 |
ENSMUST00000147521.1
ENSMUST00000167673.1 |
A630033H20Rik
|
RIKEN cDNA A630033H20 gene |
| chr7_+_55794146 | 0.90 |
ENSMUST00000032627.3
|
Tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr2_+_163602331 | 0.90 |
ENSMUST00000152135.1
|
Ttpal
|
tocopherol (alpha) transfer protein-like |
| chr1_+_136018062 | 0.90 |
ENSMUST00000117950.1
|
Tmem9
|
transmembrane protein 9 |
| chr3_-_63851251 | 0.88 |
ENSMUST00000162269.2
ENSMUST00000159676.2 ENSMUST00000175947.1 |
Plch1
|
phospholipase C, eta 1 |
| chr10_-_25200110 | 0.87 |
ENSMUST00000100012.2
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr12_+_108410542 | 0.84 |
ENSMUST00000054955.7
|
Eml1
|
echinoderm microtubule associated protein like 1 |
| chr14_-_6741430 | 0.81 |
ENSMUST00000100904.4
|
Gm3636
|
predicted gene 3636 |
| chr5_+_32247351 | 0.77 |
ENSMUST00000101376.2
|
Plb1
|
phospholipase B1 |
| chr1_-_161979636 | 0.76 |
ENSMUST00000162676.1
|
4930558K02Rik
|
RIKEN cDNA 4930558K02 gene |
| chr11_-_97782377 | 0.76 |
ENSMUST00000128801.1
|
Rpl23
|
ribosomal protein L23 |
| chr3_-_46447939 | 0.74 |
ENSMUST00000166505.1
|
Pabpc4l
|
poly(A) binding protein, cytoplasmic 4-like |
| chr11_+_31872100 | 0.74 |
ENSMUST00000020543.6
ENSMUST00000109412.2 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr6_+_83034173 | 0.74 |
ENSMUST00000000707.2
ENSMUST00000101257.3 |
Loxl3
|
lysyl oxidase-like 3 |
| chr15_+_51877742 | 0.72 |
ENSMUST00000136129.1
|
Utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr17_-_35969724 | 0.71 |
ENSMUST00000043757.8
|
Abcf1
|
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr11_-_97782409 | 0.71 |
ENSMUST00000103146.4
|
Rpl23
|
ribosomal protein L23 |
| chr17_-_24443077 | 0.71 |
ENSMUST00000119932.1
|
Dnase1l2
|
deoxyribonuclease 1-like 2 |
| chr18_+_42511496 | 0.71 |
ENSMUST00000025375.7
|
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr17_+_36898110 | 0.71 |
ENSMUST00000078438.4
|
Trim31
|
tripartite motif-containing 31 |
| chr2_+_157737401 | 0.71 |
ENSMUST00000029178.6
|
Ctnnbl1
|
catenin, beta like 1 |
| chr4_-_42034726 | 0.69 |
ENSMUST00000084677.2
|
Gm21093
|
predicted gene, 21093 |
| chr11_+_78188737 | 0.67 |
ENSMUST00000108322.2
|
Rab34
|
RAB34, member of RAS oncogene family |
| chr8_-_95294074 | 0.66 |
ENSMUST00000184103.1
|
Cngb1
|
cyclic nucleotide gated channel beta 1 |
| chr11_+_53433299 | 0.66 |
ENSMUST00000018382.6
|
Gdf9
|
growth differentiation factor 9 |
| chr4_+_105790534 | 0.65 |
ENSMUST00000185012.1
|
Gm12728
|
predicted gene 12728 |
| chr10_-_116549101 | 0.63 |
ENSMUST00000164088.1
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr2_-_132111440 | 0.62 |
ENSMUST00000128899.1
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr16_-_17722879 | 0.60 |
ENSMUST00000080936.6
ENSMUST00000012259.7 |
Med15
|
mediator complex subunit 15 |
| chr18_+_74216118 | 0.57 |
ENSMUST00000025444.6
|
Cxxc1
|
CXXC finger 1 (PHD domain) |
| chr3_-_63899437 | 0.57 |
ENSMUST00000159188.1
ENSMUST00000177143.1 |
Plch1
|
phospholipase C, eta 1 |
| chr3_-_92573715 | 0.57 |
ENSMUST00000053107.4
|
Ivl
|
involucrin |
| chr14_+_53704007 | 0.57 |
ENSMUST00000103664.4
|
Trav5-4
|
T cell receptor alpha variable 5-4 |
| chr7_+_141079125 | 0.56 |
ENSMUST00000159375.1
|
Pkp3
|
plakophilin 3 |
| chr11_-_69920892 | 0.55 |
ENSMUST00000152589.1
ENSMUST00000108612.1 ENSMUST00000108611.1 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr1_-_153186447 | 0.55 |
ENSMUST00000027753.6
|
Lamc2
|
laminin, gamma 2 |
| chr2_-_71367749 | 0.53 |
ENSMUST00000151937.1
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr2_-_76870486 | 0.52 |
ENSMUST00000138542.1
|
Ttn
|
titin |
| chr19_-_37207293 | 0.48 |
ENSMUST00000132580.1
ENSMUST00000079754.4 ENSMUST00000136286.1 ENSMUST00000126188.1 ENSMUST00000126781.1 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr2_+_163602294 | 0.47 |
ENSMUST00000171696.1
ENSMUST00000109408.3 |
Ttpal
|
tocopherol (alpha) transfer protein-like |
| chrY_-_40315451 | 0.46 |
ENSMUST00000177713.1
|
Gm21865
|
predicted gene, 21865 |
| chr2_+_25403128 | 0.45 |
ENSMUST00000154809.1
ENSMUST00000055921.7 ENSMUST00000141567.1 |
Npdc1
|
neural proliferation, differentiation and control 1 |
| chr14_+_4430992 | 0.45 |
ENSMUST00000164603.1
ENSMUST00000166848.1 |
Gm3173
|
predicted gene 3173 |
| chr7_+_43187170 | 0.43 |
ENSMUST00000072829.3
|
Zfp936
|
zinc finger protein 936 |
| chrY_-_41644045 | 0.43 |
ENSMUST00000180058.1
|
Gm21729
|
predicted gene, 21729 |
| chr10_-_12923075 | 0.43 |
ENSMUST00000180529.1
|
B230208H11Rik
|
RIKEN cDNA B230208H11 gene |
| chr11_+_100320596 | 0.42 |
ENSMUST00000152521.1
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chr4_+_43441939 | 0.42 |
ENSMUST00000060864.6
|
Tesk1
|
testis specific protein kinase 1 |
| chr1_+_110099295 | 0.39 |
ENSMUST00000134301.1
|
Cdh7
|
cadherin 7, type 2 |
| chr14_+_3225315 | 0.39 |
ENSMUST00000178670.1
|
D830030K20Rik
|
RIKEN cDNA D830030K20 gene |
| chr11_+_109413917 | 0.38 |
ENSMUST00000055404.7
|
9930022D16Rik
|
RIKEN cDNA 9930022D16 gene |
| chr9_+_46282997 | 0.38 |
ENSMUST00000074957.3
|
Bud13
|
BUD13 homolog (yeast) |
| chrY_-_31633324 | 0.38 |
ENSMUST00000179076.1
|
1700040F15Rik
|
RIKEN cDNA 1700040F15 gene |
| chr18_+_55057557 | 0.38 |
ENSMUST00000181765.1
|
Gm4221
|
predicted gene 4221 |
| chr7_+_102774495 | 0.38 |
ENSMUST00000098217.2
|
Olfr561
|
olfactory receptor 561 |
| chr18_-_36454487 | 0.36 |
ENSMUST00000025204.5
|
Pfdn1
|
prefoldin 1 |
| chr3_-_107333289 | 0.35 |
ENSMUST00000061772.9
|
Rbm15
|
RNA binding motif protein 15 |
| chr7_+_17972124 | 0.35 |
ENSMUST00000094799.2
|
Ceacam11
|
carcinoembryonic antigen-related cell adhesion molecule 11 |
| chr11_-_75796048 | 0.35 |
ENSMUST00000021209.7
|
Doc2b
|
double C2, beta |
| chr18_-_43477764 | 0.34 |
ENSMUST00000057110.9
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr2_-_80129458 | 0.33 |
ENSMUST00000102653.1
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr11_+_49247462 | 0.32 |
ENSMUST00000109194.1
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr11_-_68853019 | 0.31 |
ENSMUST00000108672.1
|
Ndel1
|
nuclear distribution gene E-like homolog 1 (A. nidulans) |
| chr12_-_84876479 | 0.31 |
ENSMUST00000163189.1
ENSMUST00000110254.2 ENSMUST00000002073.6 |
Ltbp2
|
latent transforming growth factor beta binding protein 2 |
| chr7_+_103731928 | 0.30 |
ENSMUST00000098193.1
|
Olfr628
|
olfactory receptor 628 |
| chrY_-_52470686 | 0.29 |
ENSMUST00000179424.1
|
Gm20923
|
predicted gene, 20923 |
| chr12_+_86734381 | 0.26 |
ENSMUST00000095527.5
|
Gm6772
|
predicted gene 6772 |
| chr13_-_67399738 | 0.26 |
ENSMUST00000181071.1
ENSMUST00000109732.1 |
Zfp429
|
zinc finger protein 429 |
| chr15_+_51877429 | 0.25 |
ENSMUST00000137116.2
ENSMUST00000161651.1 ENSMUST00000059599.9 |
Utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chrY_+_43953619 | 0.24 |
ENSMUST00000179551.1
|
Gm21241
|
predicted gene, 21241 |
| chr2_-_174346712 | 0.23 |
ENSMUST00000168292.1
|
Gm20721
|
predicted gene, 20721 |
| chr16_+_32400506 | 0.22 |
ENSMUST00000115149.2
|
Tm4sf19
|
transmembrane 4 L six family member 19 |
| chr7_+_103716596 | 0.20 |
ENSMUST00000098194.1
|
Olfr243
|
olfactory receptor 243 |
| chr14_-_6874257 | 0.20 |
ENSMUST00000179374.1
ENSMUST00000178298.1 |
Gm3629
Gm3667
|
predicted gene 3629 predicted gene 3667 |
| chr1_-_164935522 | 0.20 |
ENSMUST00000027860.7
|
Xcl1
|
chemokine (C motif) ligand 1 |
| chr9_+_36475580 | 0.19 |
ENSMUST00000165591.2
|
Gm9513
|
predicted gene 9513 |
| chr14_-_19569553 | 0.18 |
ENSMUST00000112595.2
|
Gm2237
|
predicted gene 2237 |
| chrY_-_29683370 | 0.16 |
ENSMUST00000180373.1
|
Gm21679
|
predicted gene, 21679 |
| chr2_-_80128834 | 0.15 |
ENSMUST00000102654.4
ENSMUST00000102655.3 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr5_+_129096740 | 0.14 |
ENSMUST00000056617.7
ENSMUST00000156437.1 |
Gpr133
|
G protein-coupled receptor 133 |
| chr1_-_174118014 | 0.14 |
ENSMUST00000063030.3
|
Olfr231
|
olfactory receptor 231 |
| chrX_-_155128409 | 0.13 |
ENSMUST00000076671.3
|
1700042B14Rik
|
RIKEN cDNA 1700042B14 gene |
| chr5_-_5479120 | 0.13 |
ENSMUST00000115447.1
|
1700015F17Rik
|
RIKEN cDNA 1700015F17 gene |
| chr15_-_85811644 | 0.12 |
ENSMUST00000144067.1
ENSMUST00000134631.1 ENSMUST00000154814.1 ENSMUST00000071876.6 ENSMUST00000150995.1 |
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chr15_-_77970750 | 0.12 |
ENSMUST00000100484.4
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chrX_-_53608979 | 0.12 |
ENSMUST00000123034.1
|
Gm14597
|
predicted gene 14597 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Neurog2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.1 | GO:0035702 | monocyte homeostasis(GO:0035702) |
| 4.0 | 12.0 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 2.9 | 17.3 | GO:0032796 | uropod organization(GO:0032796) |
| 1.9 | 7.4 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 1.8 | 9.0 | GO:1904970 | brush border assembly(GO:1904970) |
| 1.5 | 5.9 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 1.5 | 4.4 | GO:0002884 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) negative regulation of hypersensitivity(GO:0002884) |
| 1.3 | 6.5 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 1.1 | 18.4 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 1.0 | 3.0 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.9 | 3.5 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.8 | 5.8 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.7 | 2.1 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.7 | 6.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.6 | 3.2 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.6 | 3.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.5 | 14.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.5 | 3.2 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.5 | 1.0 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.4 | 1.8 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.4 | 2.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.4 | 1.6 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.4 | 2.2 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.3 | 3.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.3 | 1.7 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.3 | 1.3 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.3 | 1.0 | GO:0006404 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.3 | 4.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 1.5 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.3 | 2.6 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.2 | 0.7 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.2 | 1.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.2 | 1.1 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.2 | 0.6 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.2 | 0.2 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.2 | 1.0 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.2 | 0.9 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.2 | 1.6 | GO:0090399 | bone marrow development(GO:0048539) replicative senescence(GO:0090399) |
| 0.2 | 0.9 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.2 | 0.7 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.2 | 2.0 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.2 | 1.7 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.2 | 2.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 2.5 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.1 | 5.2 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 0.7 | GO:1905161 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.1 | 1.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.5 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.5 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.1 | 0.9 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 1.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 1.6 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 1.2 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 5.9 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.1 | 1.6 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.1 | 1.0 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 1.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 1.0 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 1.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.8 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 1.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.6 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 1.1 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.1 | 2.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 2.8 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 1.0 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 1.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.6 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.6 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 1.4 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 2.9 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 0.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 2.3 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.9 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 1.5 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 1.0 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.4 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 1.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 1.2 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 1.8 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 1.0 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.7 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.3 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.7 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 0.0 | 0.6 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 9.0 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.8 | 7.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.8 | 17.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.7 | 2.8 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.4 | 1.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.4 | 2.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.3 | 4.4 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 2.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 0.9 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.2 | 3.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 0.5 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.2 | 17.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 1.0 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 3.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 1.6 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.2 | 2.7 | GO:0044453 | nuclear membrane part(GO:0044453) |
| 0.1 | 1.8 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.6 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.1 | 6.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 1.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 6.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 2.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.7 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 5.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 2.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 12.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.0 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 3.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 4.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 2.9 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 8.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 3.3 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.0 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 9.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 18.4 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 2.2 | 6.5 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) interleukin-1 Type I receptor antagonist activity(GO:0045352) interleukin-1 Type II receptor antagonist activity(GO:0045353) |
| 1.4 | 5.8 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.9 | 3.5 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.9 | 19.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.7 | 14.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.7 | 7.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.5 | 2.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.4 | 1.8 | GO:0038100 | nodal binding(GO:0038100) |
| 0.4 | 2.1 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.3 | 2.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.3 | 12.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 2.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 0.8 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.3 | 0.8 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.2 | 0.6 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.2 | 12.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.2 | 1.5 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 1.4 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 1.0 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.7 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 1.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 16.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 2.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 3.0 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.6 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.6 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 0.9 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 4.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.6 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 1.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 2.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 4.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.5 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 0.9 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 2.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.5 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 2.7 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.0 | 0.9 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 2.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 3.1 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 3.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 3.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.7 | GO:0043855 | cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 1.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 1.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 5.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 3.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 5.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 3.2 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 1.8 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 1.5 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 11.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 1.1 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 2.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 4.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 4.4 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 18.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 8.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 2.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 6.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 2.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 3.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 3.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 9.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.5 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 12.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.5 | 8.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.3 | 11.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.3 | 16.1 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.2 | 3.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 2.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 2.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 6.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 14.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.9 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 2.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 4.5 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 3.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.8 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 2.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.0 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.1 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |