Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
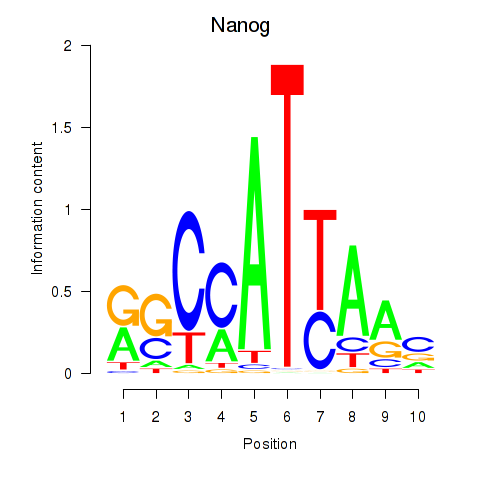
Results for Nanog
Z-value: 1.32
Transcription factors associated with Nanog
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nanog
|
ENSMUSG00000012396.6 | Nanog homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nanog | mm10_v2_chr6_+_122707489_122707608 | 0.17 | 3.2e-01 | Click! |
Activity profile of Nanog motif
Sorted Z-values of Nanog motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_41458923 | 10.52 |
ENSMUST00000031910.7
|
Prss1
|
protease, serine, 1 (trypsin 1) |
| chr19_-_11640828 | 10.49 |
ENSMUST00000112984.2
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr6_+_41392356 | 8.62 |
ENSMUST00000049079.7
|
Gm5771
|
predicted gene 5771 |
| chr7_-_142679533 | 7.48 |
ENSMUST00000162317.1
ENSMUST00000125933.1 ENSMUST00000105931.1 ENSMUST00000105930.1 ENSMUST00000105933.1 ENSMUST00000105932.1 ENSMUST00000000220.2 |
Ins2
|
insulin II |
| chr2_+_128591205 | 7.07 |
ENSMUST00000155430.1
|
Gm355
|
predicted gene 355 |
| chr17_+_29135056 | 7.06 |
ENSMUST00000087942.4
|
Rab44
|
RAB44, member RAS oncogene family |
| chr9_+_65890237 | 6.42 |
ENSMUST00000045802.6
|
2810417H13Rik
|
RIKEN cDNA 2810417H13 gene |
| chr6_+_41354105 | 6.06 |
ENSMUST00000072103.5
|
Try10
|
trypsin 10 |
| chr17_+_40811089 | 5.57 |
ENSMUST00000024721.7
|
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr1_-_75133866 | 5.54 |
ENSMUST00000027405.4
|
Slc23a3
|
solute carrier family 23 (nucleobase transporters), member 3 |
| chr14_+_27000362 | 5.24 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr12_+_109549157 | 5.21 |
ENSMUST00000128458.1
ENSMUST00000150851.1 |
Meg3
|
maternally expressed 3 |
| chr9_-_123678782 | 4.94 |
ENSMUST00000170591.1
ENSMUST00000171647.1 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chrX_+_8271133 | 4.89 |
ENSMUST00000127103.1
ENSMUST00000115591.1 |
Slc38a5
|
solute carrier family 38, member 5 |
| chr2_+_164948219 | 4.87 |
ENSMUST00000017881.2
|
Mmp9
|
matrix metallopeptidase 9 |
| chrX_+_8271381 | 4.75 |
ENSMUST00000033512.4
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr7_-_6730412 | 4.72 |
ENSMUST00000051209.4
|
Peg3
|
paternally expressed 3 |
| chr18_+_36528145 | 4.68 |
ENSMUST00000074298.6
ENSMUST00000115694.2 |
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr3_+_105870858 | 4.41 |
ENSMUST00000164730.1
|
Adora3
|
adenosine A3 receptor |
| chr6_+_41302265 | 4.17 |
ENSMUST00000031913.4
|
Try4
|
trypsin 4 |
| chr5_+_33658123 | 4.00 |
ENSMUST00000074849.6
ENSMUST00000079534.4 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chrX_+_8271642 | 3.83 |
ENSMUST00000115590.1
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr9_-_123678873 | 3.64 |
ENSMUST00000040960.6
|
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr3_+_114030532 | 3.58 |
ENSMUST00000123619.1
ENSMUST00000092155.5 |
Col11a1
|
collagen, type XI, alpha 1 |
| chr10_+_58394361 | 3.55 |
ENSMUST00000020077.4
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr7_+_79660196 | 3.48 |
ENSMUST00000035977.7
|
Ticrr
|
TOPBP1-interacting checkpoint and replication regulator |
| chr12_-_69790660 | 3.39 |
ENSMUST00000021377.4
|
Cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr16_-_18622403 | 3.38 |
ENSMUST00000167388.1
|
Gp1bb
|
glycoprotein Ib, beta polypeptide |
| chr15_+_80097866 | 3.37 |
ENSMUST00000143928.1
|
Syngr1
|
synaptogyrin 1 |
| chr5_+_33658567 | 3.36 |
ENSMUST00000114426.3
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr3_-_54915867 | 3.33 |
ENSMUST00000070342.3
|
Sertm1
|
serine rich and transmembrane domain containing 1 |
| chr3_+_88081997 | 3.32 |
ENSMUST00000071812.5
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr5_+_33658550 | 3.32 |
ENSMUST00000152847.1
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr15_-_103251465 | 3.23 |
ENSMUST00000133600.1
ENSMUST00000134554.1 ENSMUST00000156927.1 ENSMUST00000149111.1 ENSMUST00000132836.1 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr10_+_58394381 | 3.23 |
ENSMUST00000105468.1
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr3_+_105870898 | 3.09 |
ENSMUST00000010279.5
|
Adora3
|
adenosine A3 receptor |
| chr7_-_14254870 | 2.97 |
ENSMUST00000184731.1
ENSMUST00000076576.6 |
Sult2a6
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 6 |
| chr19_-_9899450 | 2.94 |
ENSMUST00000025562.7
|
Incenp
|
inner centromere protein |
| chr7_-_14123042 | 2.82 |
ENSMUST00000098809.2
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr4_-_49597425 | 2.73 |
ENSMUST00000150664.1
|
Tmem246
|
transmembrane protein 246 |
| chr9_-_20959785 | 2.71 |
ENSMUST00000177754.1
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr3_+_68869563 | 2.70 |
ENSMUST00000054551.2
|
1110032F04Rik
|
RIKEN cDNA 1110032F04 gene |
| chrX_+_134308084 | 2.66 |
ENSMUST00000081064.5
ENSMUST00000101251.1 ENSMUST00000129782.1 |
Cenpi
|
centromere protein I |
| chr11_+_44617310 | 2.55 |
ENSMUST00000081265.5
ENSMUST00000101326.3 ENSMUST00000109268.1 |
Ebf1
|
early B cell factor 1 |
| chr11_+_62248977 | 2.54 |
ENSMUST00000018644.2
|
Adora2b
|
adenosine A2b receptor |
| chr6_+_124829582 | 2.45 |
ENSMUST00000024270.7
|
Cdca3
|
cell division cycle associated 3 |
| chr12_+_102128718 | 2.40 |
ENSMUST00000159329.1
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr15_+_44457522 | 2.40 |
ENSMUST00000166957.1
ENSMUST00000038336.5 |
Pkhd1l1
|
polycystic kidney and hepatic disease 1-like 1 |
| chr7_+_19411086 | 2.39 |
ENSMUST00000003643.1
|
Ckm
|
creatine kinase, muscle |
| chr6_-_136781718 | 2.37 |
ENSMUST00000078095.6
ENSMUST00000032338.7 |
Gucy2c
|
guanylate cyclase 2c |
| chr18_-_21652362 | 2.32 |
ENSMUST00000049105.4
|
Klhl14
|
kelch-like 14 |
| chr12_+_109747903 | 2.31 |
ENSMUST00000183084.1
ENSMUST00000182300.1 |
Mirg
|
miRNA containing gene |
| chr6_-_145047725 | 2.25 |
ENSMUST00000123930.1
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr6_-_67037399 | 2.24 |
ENSMUST00000043098.6
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr2_-_116067391 | 2.23 |
ENSMUST00000140185.1
|
2700033N17Rik
|
RIKEN cDNA 2700033N17 gene |
| chr7_-_100467149 | 2.20 |
ENSMUST00000184420.1
|
RP23-308M1.2
|
RP23-308M1.2 |
| chr15_-_66831625 | 2.19 |
ENSMUST00000164163.1
|
Sla
|
src-like adaptor |
| chr7_-_133702515 | 2.19 |
ENSMUST00000153698.1
|
Uros
|
uroporphyrinogen III synthase |
| chr3_+_122044428 | 2.16 |
ENSMUST00000013995.8
|
Abca4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr6_+_124829540 | 2.12 |
ENSMUST00000150120.1
|
Cdca3
|
cell division cycle associated 3 |
| chrX_-_164250368 | 2.12 |
ENSMUST00000112263.1
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr11_-_116077606 | 2.11 |
ENSMUST00000106450.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr3_-_144760841 | 2.10 |
ENSMUST00000059091.5
|
Clca1
|
chloride channel calcium activated 1 |
| chr17_-_25727364 | 2.08 |
ENSMUST00000170070.1
ENSMUST00000048054.7 |
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 |
| chr13_-_89742244 | 2.07 |
ENSMUST00000109543.2
ENSMUST00000159337.1 ENSMUST00000159910.1 ENSMUST00000109544.2 |
Vcan
|
versican |
| chr12_-_91384403 | 2.06 |
ENSMUST00000141429.1
|
Cep128
|
centrosomal protein 128 |
| chr3_+_37312514 | 2.03 |
ENSMUST00000057975.7
ENSMUST00000108121.3 |
Bbs12
|
Bardet-Biedl syndrome 12 (human) |
| chr4_+_154960915 | 2.01 |
ENSMUST00000049621.6
|
Hes5
|
hairy and enhancer of split 5 (Drosophila) |
| chr15_-_51991679 | 2.01 |
ENSMUST00000022927.9
|
Rad21
|
RAD21 homolog (S. pombe) |
| chr3_-_116253467 | 1.99 |
ENSMUST00000090473.5
|
Gpr88
|
G-protein coupled receptor 88 |
| chr18_+_82554463 | 1.90 |
ENSMUST00000062446.7
ENSMUST00000102812.4 ENSMUST00000075372.5 ENSMUST00000080658.4 ENSMUST00000152071.1 ENSMUST00000114674.3 ENSMUST00000142850.1 ENSMUST00000133193.1 ENSMUST00000123251.1 ENSMUST00000153478.1 ENSMUST00000132369.1 |
Mbp
|
myelin basic protein |
| chr10_+_130322845 | 1.87 |
ENSMUST00000042586.8
|
Tespa1
|
thymocyte expressed, positive selection associated 1 |
| chr17_-_81649607 | 1.86 |
ENSMUST00000163680.2
ENSMUST00000086538.3 ENSMUST00000163123.1 |
Slc8a1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr6_-_102464667 | 1.86 |
ENSMUST00000032159.6
|
Cntn3
|
contactin 3 |
| chr11_+_115154139 | 1.83 |
ENSMUST00000021076.5
|
Rab37
|
RAB37, member of RAS oncogene family |
| chr3_+_105904377 | 1.81 |
ENSMUST00000000574.1
|
Adora3
|
adenosine A3 receptor |
| chr9_-_15357692 | 1.80 |
ENSMUST00000098979.3
ENSMUST00000161132.1 |
5830418K08Rik
|
RIKEN cDNA 5830418K08 gene |
| chr7_-_115846080 | 1.76 |
ENSMUST00000166207.1
|
Sox6
|
SRY-box containing gene 6 |
| chr15_+_9436028 | 1.74 |
ENSMUST00000042360.3
|
Capsl
|
calcyphosine-like |
| chr3_-_107239707 | 1.66 |
ENSMUST00000049852.8
|
Prok1
|
prokineticin 1 |
| chr6_-_50456085 | 1.64 |
ENSMUST00000146341.1
ENSMUST00000071728.4 |
Osbpl3
|
oxysterol binding protein-like 3 |
| chr13_-_19732929 | 1.60 |
ENSMUST00000151029.1
|
A530099J19Rik
|
RIKEN cDNA A530099J19 gene |
| chr6_-_136922169 | 1.56 |
ENSMUST00000032343.6
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr13_-_89742490 | 1.55 |
ENSMUST00000109546.2
|
Vcan
|
versican |
| chr12_-_83487708 | 1.54 |
ENSMUST00000177959.1
ENSMUST00000178756.1 |
Dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr11_+_62820469 | 1.54 |
ENSMUST00000108703.1
|
Trim16
|
tripartite motif-containing 16 |
| chr7_+_43437073 | 1.53 |
ENSMUST00000070518.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr17_-_51826562 | 1.51 |
ENSMUST00000024720.4
ENSMUST00000129667.1 ENSMUST00000156051.1 ENSMUST00000169480.1 ENSMUST00000148559.1 |
Satb1
|
special AT-rich sequence binding protein 1 |
| chr4_+_12906838 | 1.51 |
ENSMUST00000143186.1
ENSMUST00000183345.1 |
Triqk
|
triple QxxK/R motif containing |
| chrX_+_48146436 | 1.49 |
ENSMUST00000033427.6
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr4_+_126024506 | 1.46 |
ENSMUST00000106162.1
|
Csf3r
|
colony stimulating factor 3 receptor (granulocyte) |
| chr14_-_49245389 | 1.45 |
ENSMUST00000130853.1
ENSMUST00000022398.7 |
1700011H14Rik
|
RIKEN cDNA 1700011H14 gene |
| chr17_-_47834682 | 1.44 |
ENSMUST00000066368.6
|
Mdfi
|
MyoD family inhibitor |
| chr5_-_138172383 | 1.40 |
ENSMUST00000000505.9
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chrX_+_49470450 | 1.39 |
ENSMUST00000114904.3
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr13_+_44729794 | 1.38 |
ENSMUST00000172830.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr16_+_44765732 | 1.38 |
ENSMUST00000057488.8
|
Cd200r1
|
CD200 receptor 1 |
| chr4_+_43506966 | 1.38 |
ENSMUST00000030183.3
|
Car9
|
carbonic anhydrase 9 |
| chr7_+_28693032 | 1.36 |
ENSMUST00000151227.1
ENSMUST00000108281.1 |
Fbxo27
|
F-box protein 27 |
| chr1_-_87101590 | 1.36 |
ENSMUST00000113270.2
|
Alpi
|
alkaline phosphatase, intestinal |
| chr16_+_19760902 | 1.35 |
ENSMUST00000119468.1
|
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr12_+_69790288 | 1.34 |
ENSMUST00000021378.3
|
4930512B01Rik
|
RIKEN cDNA 4930512B01 gene |
| chr7_-_141655319 | 1.33 |
ENSMUST00000062451.7
|
Muc6
|
mucin 6, gastric |
| chr3_+_103860265 | 1.33 |
ENSMUST00000029433.7
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr6_-_145047636 | 1.33 |
ENSMUST00000149769.1
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chrX_+_49470555 | 1.31 |
ENSMUST00000042444.6
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr4_+_101507947 | 1.30 |
ENSMUST00000149047.1
ENSMUST00000106929.3 |
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr17_-_43543639 | 1.30 |
ENSMUST00000178772.1
|
Ankrd66
|
ankyrin repeat domain 66 |
| chr6_+_125552948 | 1.30 |
ENSMUST00000112254.1
ENSMUST00000112253.1 ENSMUST00000001995.7 |
Vwf
|
Von Willebrand factor homolog |
| chr13_-_47106176 | 1.29 |
ENSMUST00000021807.6
ENSMUST00000135278.1 |
Dek
|
DEK oncogene (DNA binding) |
| chr17_+_35841668 | 1.28 |
ENSMUST00000174124.1
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr18_+_35553401 | 1.26 |
ENSMUST00000181664.1
|
Snhg4
|
small nucleolar RNA host gene 4 (non-protein coding) |
| chr3_-_37312418 | 1.26 |
ENSMUST00000075537.6
ENSMUST00000071400.6 ENSMUST00000102955.4 ENSMUST00000140956.1 |
Cetn4
|
centrin 4 |
| chr2_+_57997884 | 1.26 |
ENSMUST00000112616.1
ENSMUST00000166729.1 |
Galnt5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 |
| chr13_-_12258093 | 1.25 |
ENSMUST00000099856.4
|
Mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr7_-_45694369 | 1.25 |
ENSMUST00000040636.6
|
Sec1
|
secretory blood group 1 |
| chr4_-_149454971 | 1.25 |
ENSMUST00000030848.2
|
Rbp7
|
retinol binding protein 7, cellular |
| chr11_-_20332689 | 1.24 |
ENSMUST00000109594.1
|
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr5_+_25246775 | 1.23 |
ENSMUST00000144971.1
|
Galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 |
| chr18_-_15063560 | 1.23 |
ENSMUST00000168989.1
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr3_-_49757257 | 1.22 |
ENSMUST00000035931.7
|
Pcdh18
|
protocadherin 18 |
| chr15_-_93519499 | 1.20 |
ENSMUST00000109255.2
|
Prickle1
|
prickle homolog 1 (Drosophila) |
| chr11_-_20332654 | 1.19 |
ENSMUST00000004634.6
|
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr5_+_30711849 | 1.18 |
ENSMUST00000088081.4
ENSMUST00000101442.3 |
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr6_+_125145235 | 1.17 |
ENSMUST00000119527.1
ENSMUST00000088276.6 ENSMUST00000051171.7 ENSMUST00000117675.1 |
Iffo1
|
intermediate filament family orphan 1 |
| chr2_-_45112890 | 1.14 |
ENSMUST00000076836.6
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr13_-_51567084 | 1.13 |
ENSMUST00000021898.5
|
Shc3
|
src homology 2 domain-containing transforming protein C3 |
| chr2_+_125152505 | 1.13 |
ENSMUST00000110494.2
ENSMUST00000028630.2 ENSMUST00000110495.2 |
Slc12a1
|
solute carrier family 12, member 1 |
| chr3_-_144819494 | 1.12 |
ENSMUST00000029929.7
|
Clca2
|
chloride channel calcium activated 2 |
| chr17_+_8849974 | 1.10 |
ENSMUST00000115720.1
|
Pde10a
|
phosphodiesterase 10A |
| chr19_+_8802486 | 1.09 |
ENSMUST00000172175.1
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr4_-_43046196 | 1.08 |
ENSMUST00000036462.5
|
Fam214b
|
family with sequence similarity 214, member B |
| chr13_-_19619820 | 1.08 |
ENSMUST00000002885.6
|
Epdr1
|
ependymin related protein 1 (zebrafish) |
| chr3_+_79884576 | 1.08 |
ENSMUST00000145992.1
|
Fam198b
|
family with sequence similarity 198, member B |
| chr5_-_16731074 | 1.07 |
ENSMUST00000073014.5
|
Gm8991
|
predicted pseudogene 8991 |
| chr10_-_127351753 | 1.07 |
ENSMUST00000059718.4
|
Inhbe
|
inhibin beta E |
| chr3_+_10088173 | 1.05 |
ENSMUST00000061419.7
|
Gm9833
|
predicted gene 9833 |
| chrX_+_159840463 | 1.04 |
ENSMUST00000112451.1
ENSMUST00000112453.2 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr11_-_101095367 | 1.04 |
ENSMUST00000019447.8
|
Psmc3ip
|
proteasome (prosome, macropain) 26S subunit, ATPase 3, interacting protein |
| chr1_+_172555932 | 1.03 |
ENSMUST00000061835.3
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr12_+_72441852 | 1.03 |
ENSMUST00000162159.1
|
Lrrc9
|
leucine rich repeat containing 9 |
| chr7_+_25267669 | 1.01 |
ENSMUST00000169266.1
|
Cic
|
capicua homolog (Drosophila) |
| chr19_-_32196393 | 0.98 |
ENSMUST00000151822.1
|
Sgms1
|
sphingomyelin synthase 1 |
| chr2_+_130274437 | 0.97 |
ENSMUST00000141872.1
|
Nop56
|
NOP56 ribonucleoprotein |
| chr17_+_78491549 | 0.96 |
ENSMUST00000079363.4
|
Gm10093
|
predicted pseudogene 10093 |
| chr10_+_33905015 | 0.96 |
ENSMUST00000169670.1
|
Rsph4a
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr4_-_83052229 | 0.95 |
ENSMUST00000107230.1
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr9_-_44342332 | 0.94 |
ENSMUST00000097558.3
|
Hmbs
|
hydroxymethylbilane synthase |
| chr14_-_18270953 | 0.93 |
ENSMUST00000100799.2
ENSMUST00000079419.4 ENSMUST00000080281.7 |
Rpl15
|
ribosomal protein L15 |
| chr3_-_82876483 | 0.93 |
ENSMUST00000048647.7
|
Rbm46
|
RNA binding motif protein 46 |
| chr6_-_116716888 | 0.92 |
ENSMUST00000056623.6
|
Tmem72
|
transmembrane protein 72 |
| chrX_+_7822289 | 0.92 |
ENSMUST00000009875.4
|
Kcnd1
|
potassium voltage-gated channel, Shal-related family, member 1 |
| chr8_+_110721462 | 0.91 |
ENSMUST00000052457.8
|
Mtss1l
|
metastasis suppressor 1-like |
| chr11_-_98053415 | 0.91 |
ENSMUST00000017544.2
|
Stac2
|
SH3 and cysteine rich domain 2 |
| chr15_-_13173607 | 0.91 |
ENSMUST00000036439.4
|
Cdh6
|
cadherin 6 |
| chr7_-_116084635 | 0.91 |
ENSMUST00000111755.3
|
Gm4353
|
predicted gene 4353 |
| chr15_-_34356421 | 0.90 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr3_+_65666223 | 0.89 |
ENSMUST00000099075.2
ENSMUST00000107848.1 ENSMUST00000161794.1 |
Lekr1
|
leucine, glutamate and lysine rich 1 |
| chr2_+_127909058 | 0.89 |
ENSMUST00000110344.1
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr18_+_37518341 | 0.89 |
ENSMUST00000097609.1
|
Pcdhb22
|
protocadherin beta 22 |
| chr5_-_99978914 | 0.89 |
ENSMUST00000112939.3
ENSMUST00000171786.1 ENSMUST00000072750.6 ENSMUST00000019128.8 ENSMUST00000172361.1 |
Hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr4_+_101507855 | 0.87 |
ENSMUST00000038207.5
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr1_-_172027251 | 0.85 |
ENSMUST00000138714.1
|
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chr17_+_7170101 | 0.85 |
ENSMUST00000024575.6
|
Rps6ka2
|
ribosomal protein S6 kinase, polypeptide 2 |
| chr6_-_142473075 | 0.85 |
ENSMUST00000032371.7
|
Gys2
|
glycogen synthase 2 |
| chr3_-_55055038 | 0.84 |
ENSMUST00000029368.2
|
Ccna1
|
cyclin A1 |
| chr7_+_113514085 | 0.81 |
ENSMUST00000122890.1
|
Far1
|
fatty acyl CoA reductase 1 |
| chr16_+_20696175 | 0.80 |
ENSMUST00000128273.1
|
Fam131a
|
family with sequence similarity 131, member A |
| chr7_+_128246953 | 0.79 |
ENSMUST00000167965.1
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr9_+_106368594 | 0.79 |
ENSMUST00000172306.2
|
Dusp7
|
dual specificity phosphatase 7 |
| chr4_+_48585193 | 0.79 |
ENSMUST00000107703.1
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr11_-_40733373 | 0.78 |
ENSMUST00000020579.8
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr7_+_112742025 | 0.77 |
ENSMUST00000164363.1
|
Tead1
|
TEA domain family member 1 |
| chr6_+_71909046 | 0.76 |
ENSMUST00000055296.8
|
Polr1a
|
polymerase (RNA) I polypeptide A |
| chr10_-_61476937 | 0.76 |
ENSMUST00000051330.4
|
D830039M14Rik
|
RIKEN cDNA D830039M14 gene |
| chr6_-_29179584 | 0.76 |
ENSMUST00000159200.1
|
Prrt4
|
proline-rich transmembrane protein 4 |
| chr8_-_120589304 | 0.76 |
ENSMUST00000034278.5
|
Gins2
|
GINS complex subunit 2 (Psf2 homolog) |
| chr3_+_54481429 | 0.75 |
ENSMUST00000091130.3
|
Gm5641
|
predicted gene 5641 |
| chr11_-_99244058 | 0.73 |
ENSMUST00000103132.3
ENSMUST00000038214.6 |
Krt222
|
keratin 222 |
| chr13_+_23752267 | 0.72 |
ENSMUST00000091703.2
|
Hist1h3b
|
histone cluster 1, H3b |
| chr10_+_62252325 | 0.72 |
ENSMUST00000020278.5
|
Tacr2
|
tachykinin receptor 2 |
| chr12_+_72441933 | 0.71 |
ENSMUST00000161284.1
|
Lrrc9
|
leucine rich repeat containing 9 |
| chr7_-_110769345 | 0.71 |
ENSMUST00000098108.2
|
B430319F04Rik
|
RIKEN cDNA B430319F04 gene |
| chrX_-_56822308 | 0.71 |
ENSMUST00000135542.1
ENSMUST00000114766.1 |
Mtap7d3
|
MAP7 domain containing 3 |
| chr1_+_89454769 | 0.69 |
ENSMUST00000027521.8
ENSMUST00000074945.5 |
Agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr10_-_62422427 | 0.68 |
ENSMUST00000020277.8
|
Hkdc1
|
hexokinase domain containing 1 |
| chr6_-_124779686 | 0.67 |
ENSMUST00000147669.1
ENSMUST00000128697.1 ENSMUST00000032218.3 ENSMUST00000112475.2 |
Lrrc23
|
leucine rich repeat containing 23 |
| chr13_+_111686303 | 0.67 |
ENSMUST00000047412.4
ENSMUST00000109271.2 |
Mier3
|
mesoderm induction early response 1, family member 3 |
| chrX_+_169879596 | 0.64 |
ENSMUST00000112105.1
ENSMUST00000078947.5 |
Mid1
|
midline 1 |
| chr4_+_109415631 | 0.64 |
ENSMUST00000106618.1
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr2_+_112379204 | 0.63 |
ENSMUST00000028552.3
|
Katnbl1
|
katanin p80 subunit B like 1 |
| chr4_-_11386757 | 0.62 |
ENSMUST00000108313.1
ENSMUST00000108311.2 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr3_-_63851251 | 0.62 |
ENSMUST00000162269.2
ENSMUST00000159676.2 ENSMUST00000175947.1 |
Plch1
|
phospholipase C, eta 1 |
| chr3_+_37348645 | 0.62 |
ENSMUST00000038885.3
|
Fgf2
|
fibroblast growth factor 2 |
| chr12_+_3891728 | 0.61 |
ENSMUST00000172689.1
ENSMUST00000111186.1 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr7_-_105574324 | 0.61 |
ENSMUST00000081165.7
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr19_+_25672408 | 0.61 |
ENSMUST00000053068.5
|
Dmrt2
|
doublesex and mab-3 related transcription factor 2 |
| chr7_+_128246812 | 0.61 |
ENSMUST00000164710.1
ENSMUST00000070656.5 |
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr1_+_135584773 | 0.60 |
ENSMUST00000067468.4
|
Gm4793
|
predicted gene 4793 |
| chr17_-_32800938 | 0.60 |
ENSMUST00000080905.6
|
Zfp811
|
zinc finger protein 811 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nanog
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) neuron projection maintenance(GO:1990535) |
| 2.1 | 8.6 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 1.9 | 5.6 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 1.6 | 4.9 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.9 | 9.3 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.9 | 13.5 | GO:0015816 | glycine transport(GO:0015816) |
| 0.8 | 6.8 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.7 | 0.7 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.7 | 2.7 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.7 | 2.0 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.6 | 1.9 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.6 | 10.7 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.6 | 1.9 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.6 | 2.4 | GO:0015824 | proline transport(GO:0015824) L-serine transport(GO:0015825) |
| 0.6 | 1.2 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.6 | 3.6 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.5 | 2.2 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.5 | 5.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.5 | 3.6 | GO:0035989 | tendon development(GO:0035989) |
| 0.5 | 2.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.4 | 1.3 | GO:0071661 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 0.4 | 3.5 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.4 | 2.5 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.4 | 1.9 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.4 | 2.2 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.3 | 0.6 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.3 | 0.9 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.3 | 2.0 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.3 | 0.9 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.3 | 3.2 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.2 | 2.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.2 | 2.1 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 1.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.2 | 0.6 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.2 | 0.6 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.2 | 28.2 | GO:0007586 | digestion(GO:0007586) |
| 0.2 | 0.8 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.2 | 1.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 2.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 1.3 | GO:0044838 | cell quiescence(GO:0044838) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.2 | 4.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 0.5 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) chromosome movement towards spindle pole(GO:0051305) formin-nucleated actin cable assembly(GO:0070649) |
| 0.2 | 1.8 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 5.2 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.2 | 1.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 2.7 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.2 | 1.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 0.5 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.2 | 0.6 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 1.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 1.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.3 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.1 | 0.8 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.5 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 2.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.8 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 2.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 1.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 2.4 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 1.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.3 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 1.5 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 | 0.9 | GO:0045835 | brain renin-angiotensin system(GO:0002035) negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 0.9 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.1 | 0.6 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.4 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 1.0 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.3 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002587) |
| 0.1 | 3.9 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 1.5 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.3 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 | 1.4 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 1.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.4 | GO:0006363 | transcription initiation from RNA polymerase I promoter(GO:0006361) termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 2.1 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.1 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 1.9 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 2.4 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 0.8 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.4 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 1.0 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.5 | GO:0072658 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 1.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 1.8 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 2.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.4 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 2.4 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.6 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.9 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.3 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.2 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.3 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 2.9 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.1 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.0 | 0.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 1.1 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.0 | 0.1 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) |
| 0.0 | 1.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.4 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 1.0 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 1.2 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.8 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.2 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 1.9 | GO:0071773 | response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.0 | 0.3 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) uterus morphogenesis(GO:0061038) |
| 0.0 | 0.2 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.4 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 1.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.4 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.3 | 3.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 2.9 | GO:0000801 | central element(GO:0000801) |
| 0.3 | 2.0 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.3 | 3.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 0.8 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 1.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 2.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 0.9 | GO:0060187 | cell pole(GO:0060187) |
| 0.2 | 0.8 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 2.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 2.0 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.6 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 0.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 0.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 1.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 2.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 2.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.4 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 1.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 4.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 2.1 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.9 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 3.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 3.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 4.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 1.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 38.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 4.1 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 2.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 3.9 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 6.4 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.9 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.1 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 5.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 13.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 1.1 | 11.8 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 1.0 | 11.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.8 | 5.8 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.6 | 3.6 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.5 | 3.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.5 | 3.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.5 | 1.9 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.5 | 1.4 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.4 | 1.3 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.4 | 3.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.4 | 2.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.4 | 2.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.4 | 1.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.3 | 5.6 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.3 | 0.9 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.3 | 0.8 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.2 | 0.7 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 4.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 0.7 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.2 | 1.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 0.6 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.2 | 1.0 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 1.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 2.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 0.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 4.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.3 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.2 | 6.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 1.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 3.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 0.8 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 1.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 1.5 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.2 | 31.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 2.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 2.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.3 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.1 | 0.6 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.1 | 3.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.2 | GO:0019841 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.1 | 1.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.9 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.7 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.4 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 2.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.5 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 1.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 3.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 2.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 0.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 3.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0052740 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.8 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.6 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 1.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 3.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 5.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.2 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 1.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 8.4 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 2.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 12.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.3 | 0.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 8.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 6.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 3.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 5.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 5.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 2.0 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 2.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 2.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 7.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 2.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 3.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.6 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.5 | 15.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 16.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 4.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 6.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 3.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 6.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.2 | 1.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 3.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 2.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 9.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.9 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 5.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 7.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.7 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 0.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 0.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 2.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 0.9 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 2.6 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.6 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 1.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 2.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 2.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 3.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |