Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
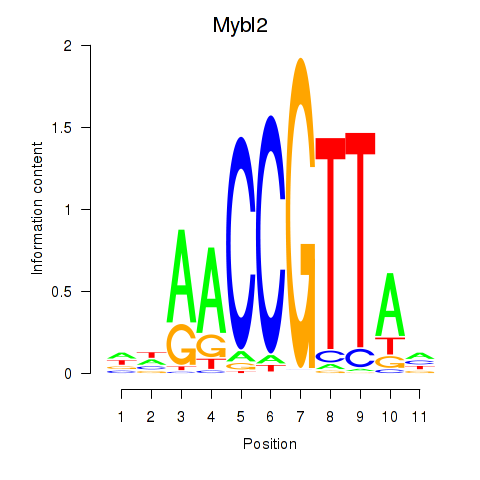
Results for Mybl2
Z-value: 1.71
Transcription factors associated with Mybl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mybl2
|
ENSMUSG00000017861.5 | myeloblastosis oncogene-like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mybl2 | mm10_v2_chr2_+_163054682_163054693 | 0.89 | 5.7e-13 | Click! |
Activity profile of Mybl2 motif
Sorted Z-values of Mybl2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_86078070 | 15.68 |
ENSMUST00000032069.5
|
Add2
|
adducin 2 (beta) |
| chr7_-_24760311 | 13.87 |
ENSMUST00000063956.5
|
Cd177
|
CD177 antigen |
| chr3_+_51661167 | 13.07 |
ENSMUST00000099106.3
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr11_-_83286722 | 11.99 |
ENSMUST00000163961.2
|
Slfn14
|
schlafen family member 14 |
| chr1_-_132390301 | 11.87 |
ENSMUST00000132435.1
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr19_-_41802028 | 10.71 |
ENSMUST00000026150.8
ENSMUST00000177495.1 ENSMUST00000163265.1 |
Arhgap19
|
Rho GTPase activating protein 19 |
| chr2_+_84988194 | 9.55 |
ENSMUST00000028466.5
|
Prg3
|
proteoglycan 3 |
| chr1_-_169531343 | 9.52 |
ENSMUST00000028000.7
|
Nuf2
|
NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr5_-_24030297 | 8.98 |
ENSMUST00000101513.2
|
Fam126a
|
family with sequence similarity 126, member A |
| chr7_-_127137807 | 8.58 |
ENSMUST00000049931.5
|
Spn
|
sialophorin |
| chr2_+_25372315 | 8.08 |
ENSMUST00000028329.6
ENSMUST00000114293.2 ENSMUST00000100323.2 |
Sapcd2
|
suppressor APC domain containing 2 |
| chr4_-_124936852 | 7.85 |
ENSMUST00000030690.5
ENSMUST00000084296.3 |
Cdca8
|
cell division cycle associated 8 |
| chr1_+_40439627 | 7.79 |
ENSMUST00000097772.3
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr1_-_169531447 | 7.71 |
ENSMUST00000111368.1
|
Nuf2
|
NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr3_+_51661209 | 7.47 |
ENSMUST00000161590.1
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr3_-_36571952 | 7.27 |
ENSMUST00000029270.3
|
Ccna2
|
cyclin A2 |
| chr19_-_15924560 | 7.17 |
ENSMUST00000162053.1
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr6_+_113531675 | 7.13 |
ENSMUST00000036340.5
ENSMUST00000101051.2 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr18_+_34624621 | 6.94 |
ENSMUST00000167161.1
|
Kif20a
|
kinesin family member 20A |
| chr1_+_57995971 | 6.89 |
ENSMUST00000027202.8
|
Sgol2
|
shugoshin-like 2 (S. pombe) |
| chr2_+_29124106 | 6.85 |
ENSMUST00000129544.1
|
Setx
|
senataxin |
| chr14_-_67715585 | 6.79 |
ENSMUST00000163100.1
ENSMUST00000132705.1 ENSMUST00000124045.1 |
Cdca2
|
cell division cycle associated 2 |
| chr11_-_116076986 | 6.66 |
ENSMUST00000153408.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr13_-_23551648 | 6.19 |
ENSMUST00000102971.1
|
Hist1h4f
|
histone cluster 1, H4f |
| chr4_+_49059256 | 6.14 |
ENSMUST00000076670.2
|
E130309F12Rik
|
RIKEN cDNA E130309F12 gene |
| chr7_-_4812351 | 5.82 |
ENSMUST00000079496.7
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chrX_-_143827391 | 5.68 |
ENSMUST00000087316.5
|
Capn6
|
calpain 6 |
| chr8_+_117498272 | 5.62 |
ENSMUST00000081232.7
|
Plcg2
|
phospholipase C, gamma 2 |
| chr6_-_122609964 | 5.43 |
ENSMUST00000032211.4
|
Gdf3
|
growth differentiation factor 3 |
| chr15_-_55090422 | 5.39 |
ENSMUST00000110231.1
ENSMUST00000023059.6 |
Dscc1
|
defective in sister chromatid cohesion 1 homolog (S. cerevisiae) |
| chr9_+_65890237 | 5.33 |
ENSMUST00000045802.6
|
2810417H13Rik
|
RIKEN cDNA 2810417H13 gene |
| chr19_-_5726240 | 5.24 |
ENSMUST00000049295.8
ENSMUST00000075606.4 |
Ehbp1l1
|
EH domain binding protein 1-like 1 |
| chr14_-_87141206 | 5.17 |
ENSMUST00000022599.7
|
Diap3
|
diaphanous homolog 3 (Drosophila) |
| chr6_-_39419967 | 5.01 |
ENSMUST00000122996.1
|
Mkrn1
|
makorin, ring finger protein, 1 |
| chr11_-_116077562 | 5.00 |
ENSMUST00000174822.1
|
Unc13d
|
unc-13 homolog D (C. elegans) |
| chr8_-_92355764 | 4.98 |
ENSMUST00000180102.1
ENSMUST00000179421.1 ENSMUST00000179222.1 ENSMUST00000179029.1 |
4933436C20Rik
|
RIKEN cDNA 4933436C20 gene |
| chr11_+_58948890 | 4.87 |
ENSMUST00000078267.3
|
Hist3h2ba
|
histone cluster 3, H2ba |
| chr13_-_59675754 | 4.80 |
ENSMUST00000022039.5
ENSMUST00000095739.2 |
Golm1
|
golgi membrane protein 1 |
| chr6_-_39419757 | 4.78 |
ENSMUST00000146785.1
ENSMUST00000114823.1 |
Mkrn1
|
makorin, ring finger protein, 1 |
| chr18_+_34625009 | 4.74 |
ENSMUST00000166044.1
|
Kif20a
|
kinesin family member 20A |
| chr2_+_13573927 | 4.63 |
ENSMUST00000141365.1
ENSMUST00000028062.2 |
Vim
|
vimentin |
| chr3_+_108739658 | 4.56 |
ENSMUST00000133931.2
|
Aknad1
|
AKNA domain containing 1 |
| chr5_+_108132885 | 4.55 |
ENSMUST00000047677.7
|
Ccdc18
|
coiled-coil domain containing 18 |
| chr12_-_78906929 | 4.51 |
ENSMUST00000021544.7
|
Plek2
|
pleckstrin 2 |
| chr1_-_191575534 | 4.50 |
ENSMUST00000027933.5
|
Dtl
|
denticleless homolog (Drosophila) |
| chr15_+_79891631 | 4.27 |
ENSMUST00000177350.1
ENSMUST00000177483.1 |
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr11_-_4594750 | 4.21 |
ENSMUST00000109943.3
|
Mtmr3
|
myotubularin related protein 3 |
| chr14_-_89898466 | 4.04 |
ENSMUST00000081204.4
|
Gm10110
|
predicted gene 10110 |
| chr7_-_30280335 | 4.03 |
ENSMUST00000108190.1
|
Wdr62
|
WD repeat domain 62 |
| chrX_+_150547375 | 4.03 |
ENSMUST00000066337.6
ENSMUST00000112715.1 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr16_-_4003750 | 3.93 |
ENSMUST00000171658.1
ENSMUST00000171762.1 |
Slx4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) |
| chr4_-_116627478 | 3.84 |
ENSMUST00000081182.4
ENSMUST00000030457.5 |
Nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr13_-_49652714 | 3.82 |
ENSMUST00000021818.7
|
Cenpp
|
centromere protein P |
| chr1_+_191821444 | 3.82 |
ENSMUST00000027931.7
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2 |
| chr4_-_116627921 | 3.82 |
ENSMUST00000030456.7
|
Nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr11_+_94990996 | 3.76 |
ENSMUST00000038696.5
|
Ppp1r9b
|
protein phosphatase 1, regulatory subunit 9B |
| chr17_+_8924109 | 3.74 |
ENSMUST00000149440.1
|
Pde10a
|
phosphodiesterase 10A |
| chr5_+_34949435 | 3.68 |
ENSMUST00000030984.7
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr11_+_106751226 | 3.51 |
ENSMUST00000147326.2
ENSMUST00000182896.1 ENSMUST00000182908.1 ENSMUST00000086353.4 |
Milr1
|
mast cell immunoglobulin like receptor 1 |
| chr18_+_67800101 | 3.51 |
ENSMUST00000025425.5
|
Cep192
|
centrosomal protein 192 |
| chr14_+_44102654 | 3.41 |
ENSMUST00000074839.6
|
Ear2
|
eosinophil-associated, ribonuclease A family, member 2 |
| chr10_-_85102487 | 3.39 |
ENSMUST00000059383.6
|
Fhl4
|
four and a half LIM domains 4 |
| chr8_+_83715504 | 3.36 |
ENSMUST00000109810.1
|
Ddx39
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 |
| chr8_-_80739497 | 3.35 |
ENSMUST00000043359.8
|
Smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr2_-_18998126 | 3.34 |
ENSMUST00000006912.5
|
Pip4k2a
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chr4_+_99829437 | 3.33 |
ENSMUST00000124547.1
ENSMUST00000106994.1 |
Efcab7
|
EF-hand calcium binding domain 7 |
| chr11_+_78346666 | 3.27 |
ENSMUST00000100755.3
|
Unc119
|
unc-119 homolog (C. elegans) |
| chr13_-_4150628 | 3.27 |
ENSMUST00000110704.2
ENSMUST00000021635.7 |
Akr1c18
|
aldo-keto reductase family 1, member C18 |
| chr4_+_135626655 | 3.25 |
ENSMUST00000153347.1
|
1700029M20Rik
|
RIKEN cDNA 1700029M20 gene |
| chr9_+_75071579 | 3.20 |
ENSMUST00000136731.1
|
Myo5a
|
myosin VA |
| chr14_-_43923368 | 3.15 |
ENSMUST00000163652.1
|
Ear10
|
eosinophil-associated, ribonuclease A family, member 10 |
| chr8_+_105827721 | 3.13 |
ENSMUST00000034365.4
|
Tsnaxip1
|
translin-associated factor X (Tsnax) interacting protein 1 |
| chr12_-_112829351 | 3.12 |
ENSMUST00000062092.5
|
Cdca4
|
cell division cycle associated 4 |
| chr11_+_106276715 | 3.09 |
ENSMUST00000044462.3
|
Tcam1
|
testicular cell adhesion molecule 1 |
| chr8_+_83608175 | 3.09 |
ENSMUST00000005620.8
|
Dnajb1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr4_+_130055010 | 3.08 |
ENSMUST00000123617.1
|
Col16a1
|
collagen, type XVI, alpha 1 |
| chr12_+_111406809 | 3.04 |
ENSMUST00000150384.1
|
A230065H16Rik
|
RIKEN cDNA A230065H16 gene |
| chr8_+_57488053 | 3.04 |
ENSMUST00000180690.1
|
2500002B13Rik
|
RIKEN cDNA 2500002B13 gene |
| chr9_-_95750335 | 3.03 |
ENSMUST00000053785.3
|
Trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr11_+_106751255 | 2.99 |
ENSMUST00000183111.1
ENSMUST00000106794.2 |
Milr1
|
mast cell immunoglobulin like receptor 1 |
| chr5_+_76588663 | 2.97 |
ENSMUST00000121979.1
|
Cep135
|
centrosomal protein 135 |
| chr2_+_18998332 | 2.96 |
ENSMUST00000028069.1
|
4930426L09Rik
|
RIKEN cDNA 4930426L09 gene |
| chr12_+_106010263 | 2.92 |
ENSMUST00000021539.8
ENSMUST00000085026.4 ENSMUST00000072040.5 |
Vrk1
|
vaccinia related kinase 1 |
| chr13_-_55528511 | 2.91 |
ENSMUST00000047877.4
|
Dok3
|
docking protein 3 |
| chr19_+_33822908 | 2.87 |
ENSMUST00000042061.6
|
Gm5519
|
predicted pseudogene 5519 |
| chr10_+_75954514 | 2.85 |
ENSMUST00000099577.3
|
Gm5134
|
predicted gene 5134 |
| chr17_-_33718591 | 2.85 |
ENSMUST00000174040.1
ENSMUST00000173015.1 ENSMUST00000066121.6 ENSMUST00000172767.1 ENSMUST00000173329.1 |
March2
|
membrane-associated ring finger (C3HC4) 2 |
| chr10_+_128909866 | 2.82 |
ENSMUST00000026407.7
|
Cd63
|
CD63 antigen |
| chr7_+_19577287 | 2.79 |
ENSMUST00000108453.1
|
Zfp296
|
zinc finger protein 296 |
| chr14_-_54686060 | 2.78 |
ENSMUST00000125265.1
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr7_+_30280094 | 2.69 |
ENSMUST00000108187.1
ENSMUST00000014072.5 |
Thap8
|
THAP domain containing 8 |
| chr7_+_46861203 | 2.68 |
ENSMUST00000014545.4
|
Ldhc
|
lactate dehydrogenase C |
| chr11_+_29373618 | 2.66 |
ENSMUST00000040182.6
ENSMUST00000109477.1 |
Ccdc88a
|
coiled coil domain containing 88A |
| chr11_-_61342821 | 2.66 |
ENSMUST00000134423.1
ENSMUST00000093029.2 |
Slc47a2
|
solute carrier family 47, member 2 |
| chr19_+_34192229 | 2.62 |
ENSMUST00000054956.8
|
Stambpl1
|
STAM binding protein like 1 |
| chr11_+_69965396 | 2.62 |
ENSMUST00000018713.6
|
Cldn7
|
claudin 7 |
| chr15_+_103240405 | 2.59 |
ENSMUST00000036004.9
ENSMUST00000087351.7 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr4_+_140906344 | 2.57 |
ENSMUST00000030765.6
|
Padi2
|
peptidyl arginine deiminase, type II |
| chr4_-_126325672 | 2.57 |
ENSMUST00000102616.1
|
Tekt2
|
tektin 2 |
| chr1_-_175688353 | 2.54 |
ENSMUST00000104984.1
|
Chml
|
choroideremia-like |
| chr13_-_47105790 | 2.53 |
ENSMUST00000129352.1
|
Dek
|
DEK oncogene (DNA binding) |
| chr4_+_100776664 | 2.47 |
ENSMUST00000030257.8
ENSMUST00000097955.2 |
Cachd1
|
cache domain containing 1 |
| chr17_+_8849974 | 2.46 |
ENSMUST00000115720.1
|
Pde10a
|
phosphodiesterase 10A |
| chr7_-_10495322 | 2.42 |
ENSMUST00000032551.7
|
Zik1
|
zinc finger protein interacting with K protein 1 |
| chr17_+_35841668 | 2.41 |
ENSMUST00000174124.1
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr5_-_149051300 | 2.39 |
ENSMUST00000110505.1
|
Hmgb1
|
high mobility group box 1 |
| chr2_-_73452666 | 2.37 |
ENSMUST00000151939.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr8_+_83715177 | 2.37 |
ENSMUST00000019576.8
|
Ddx39
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 |
| chr9_+_107554633 | 2.36 |
ENSMUST00000010211.4
|
Rassf1
|
Ras association (RalGDS/AF-6) domain family member 1 |
| chr7_+_25627604 | 2.35 |
ENSMUST00000076034.6
|
B3gnt8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
| chr9_-_97111117 | 2.35 |
ENSMUST00000085206.4
|
Slc25a36
|
solute carrier family 25, member 36 |
| chr4_+_154237525 | 2.32 |
ENSMUST00000152159.1
|
Megf6
|
multiple EGF-like-domains 6 |
| chr5_-_112251137 | 2.31 |
ENSMUST00000112383.1
|
Cryba4
|
crystallin, beta A4 |
| chr10_+_58446845 | 2.31 |
ENSMUST00000003310.5
|
Ranbp2
|
RAN binding protein 2 |
| chr14_+_65598546 | 2.27 |
ENSMUST00000150897.1
|
Nuggc
|
nuclear GTPase, germinal center associated |
| chr8_+_83715239 | 2.26 |
ENSMUST00000172396.1
|
Ddx39
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 |
| chr19_-_6128208 | 2.24 |
ENSMUST00000025702.7
|
Snx15
|
sorting nexin 15 |
| chr19_-_47919269 | 2.21 |
ENSMUST00000095998.5
|
Itprip
|
inositol 1,4,5-triphosphate receptor interacting protein |
| chr19_-_8798495 | 2.20 |
ENSMUST00000096261.3
|
Polr2g
|
polymerase (RNA) II (DNA directed) polypeptide G |
| chr2_-_35336969 | 2.14 |
ENSMUST00000028241.6
|
Stom
|
stomatin |
| chrX_-_57338598 | 2.13 |
ENSMUST00000033468.4
ENSMUST00000114736.1 |
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr2_-_73453918 | 2.12 |
ENSMUST00000102679.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr3_+_90266507 | 2.07 |
ENSMUST00000098914.3
|
Dennd4b
|
DENN/MADD domain containing 4B |
| chr4_+_127021311 | 2.07 |
ENSMUST00000030623.7
|
Sfpq
|
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
| chr3_-_69004475 | 2.05 |
ENSMUST00000154741.1
ENSMUST00000148031.1 |
Ift80
|
intraflagellar transport 80 |
| chr9_+_108826320 | 2.04 |
ENSMUST00000024238.5
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) |
| chr10_+_85102627 | 2.01 |
ENSMUST00000095383.4
|
AI597468
|
expressed sequence AI597468 |
| chr12_+_119260930 | 1.99 |
ENSMUST00000022467.9
|
Gm6768
|
predicted gene 6768 |
| chr5_+_114774677 | 1.98 |
ENSMUST00000102578.4
|
Ankrd13a
|
ankyrin repeat domain 13a |
| chrX_-_157492280 | 1.98 |
ENSMUST00000112529.1
|
Sms
|
spermine synthase |
| chr8_-_40634750 | 1.96 |
ENSMUST00000173957.1
|
Mtmr7
|
myotubularin related protein 7 |
| chr11_-_98729374 | 1.96 |
ENSMUST00000126565.1
ENSMUST00000100500.2 ENSMUST00000017354.6 |
Med24
|
mediator complex subunit 24 |
| chr9_+_14784638 | 1.95 |
ENSMUST00000034405.4
|
Mre11a
|
meiotic recombination 11 homolog A (S. cerevisiae) |
| chr17_+_34398802 | 1.95 |
ENSMUST00000114175.1
ENSMUST00000078615.5 ENSMUST00000139063.1 ENSMUST00000097348.2 |
BC051142
|
cDNA sequence BC051142 |
| chr10_-_100589205 | 1.94 |
ENSMUST00000054471.8
|
4930430F08Rik
|
RIKEN cDNA 4930430F08 gene |
| chr2_+_121506748 | 1.92 |
ENSMUST00000099473.3
ENSMUST00000110602.2 |
Wdr76
|
WD repeat domain 76 |
| chr12_+_99884498 | 1.89 |
ENSMUST00000153627.1
|
Tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr7_-_28598140 | 1.89 |
ENSMUST00000040531.8
ENSMUST00000108283.1 |
Samd4b
Pak4
|
sterile alpha motif domain containing 4B p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr15_-_36608959 | 1.88 |
ENSMUST00000001809.8
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr4_+_62525369 | 1.86 |
ENSMUST00000062145.1
|
4933430I17Rik
|
RIKEN cDNA 4933430I17 gene |
| chr2_+_155956537 | 1.85 |
ENSMUST00000109619.2
ENSMUST00000039994.7 ENSMUST00000094421.4 ENSMUST00000151569.1 ENSMUST00000109618.1 |
Cep250
|
centrosomal protein 250 |
| chr11_+_3332426 | 1.80 |
ENSMUST00000136474.1
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr17_+_5492558 | 1.80 |
ENSMUST00000089185.4
|
Zdhhc14
|
zinc finger, DHHC domain containing 14 |
| chr4_-_107810948 | 1.76 |
ENSMUST00000097930.1
|
B230314M03Rik
|
RIKEN cDNA B230314M03 gene |
| chr9_+_14784660 | 1.74 |
ENSMUST00000115632.3
ENSMUST00000147305.1 |
Mre11a
|
meiotic recombination 11 homolog A (S. cerevisiae) |
| chr14_+_75284343 | 1.67 |
ENSMUST00000022577.5
|
Zc3h13
|
zinc finger CCCH type containing 13 |
| chr19_-_6128144 | 1.66 |
ENSMUST00000154601.1
ENSMUST00000138931.1 |
Snx15
|
sorting nexin 15 |
| chr6_-_47594967 | 1.64 |
ENSMUST00000081721.6
ENSMUST00000114618.1 ENSMUST00000114616.1 |
Ezh2
|
enhancer of zeste homolog 2 (Drosophila) |
| chrX_-_139714481 | 1.64 |
ENSMUST00000183728.1
|
Gm15013
|
predicted gene 15013 |
| chr6_+_125145235 | 1.63 |
ENSMUST00000119527.1
ENSMUST00000088276.6 ENSMUST00000051171.7 ENSMUST00000117675.1 |
Iffo1
|
intermediate filament family orphan 1 |
| chr7_+_25681158 | 1.61 |
ENSMUST00000108403.3
|
B9d2
|
B9 protein domain 2 |
| chr3_+_116594959 | 1.61 |
ENSMUST00000029571.8
|
Sass6
|
spindle assembly 6 homolog (C. elegans) |
| chrX_+_138914422 | 1.61 |
ENSMUST00000064937.7
ENSMUST00000113052.1 |
Nrk
|
Nik related kinase |
| chr13_+_51645232 | 1.60 |
ENSMUST00000075853.5
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr15_-_80083374 | 1.59 |
ENSMUST00000081650.7
|
Rpl3
|
ribosomal protein L3 |
| chr9_+_96119362 | 1.58 |
ENSMUST00000085217.5
ENSMUST00000122383.1 |
Gk5
|
glycerol kinase 5 (putative) |
| chr3_-_130730375 | 1.55 |
ENSMUST00000079085.6
|
Rpl34
|
ribosomal protein L34 |
| chr7_+_121083322 | 1.54 |
ENSMUST00000047025.8
ENSMUST00000170106.1 |
Otoa
|
otoancorin |
| chr7_+_102065713 | 1.52 |
ENSMUST00000094129.2
ENSMUST00000094130.2 ENSMUST00000084843.3 |
Trpc2
|
transient receptor potential cation channel, subfamily C, member 2 |
| chr18_+_14706145 | 1.50 |
ENSMUST00000040860.1
|
Psma8
|
proteasome (prosome, macropain) subunit, alpha type, 8 |
| chr19_+_53142756 | 1.50 |
ENSMUST00000050096.7
|
Add3
|
adducin 3 (gamma) |
| chr14_+_67716095 | 1.49 |
ENSMUST00000078053.6
ENSMUST00000145542.1 ENSMUST00000125212.1 |
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr3_-_95995698 | 1.46 |
ENSMUST00000130043.1
|
Plekho1
|
pleckstrin homology domain containing, family O member 1 |
| chr6_-_113600645 | 1.45 |
ENSMUST00000035870.4
|
Fancd2os
|
Fancd2 opposite strand |
| chrY_+_90755657 | 1.44 |
ENSMUST00000167967.2
|
Gm21857
|
predicted gene, 21857 |
| chr4_-_136021722 | 1.44 |
ENSMUST00000030427.5
|
Tceb3
|
transcription elongation factor B (SIII), polypeptide 3 |
| chr6_+_57702601 | 1.43 |
ENSMUST00000072954.1
ENSMUST00000050077.8 |
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr7_-_35537677 | 1.43 |
ENSMUST00000127472.1
ENSMUST00000032701.7 |
Tdrd12
|
tudor domain containing 12 |
| chr8_-_94037007 | 1.43 |
ENSMUST00000034204.9
|
Nudt21
|
nudix (nucleoside diphosphate linked moiety X)-type motif 21 |
| chr3_-_69004565 | 1.42 |
ENSMUST00000169064.1
|
Ift80
|
intraflagellar transport 80 |
| chr9_-_45984816 | 1.42 |
ENSMUST00000172450.1
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr19_-_53464721 | 1.41 |
ENSMUST00000180489.1
|
5830416P10Rik
|
RIKEN cDNA 5830416P10 gene |
| chr10_+_69534039 | 1.41 |
ENSMUST00000182557.1
|
Ank3
|
ankyrin 3, epithelial |
| chr7_+_46847128 | 1.40 |
ENSMUST00000005051.4
|
Ldha
|
lactate dehydrogenase A |
| chr4_-_88676924 | 1.39 |
ENSMUST00000105148.1
|
Gm13280
|
predicted gene 13280 |
| chr3_-_130730310 | 1.35 |
ENSMUST00000062601.7
|
Rpl34
|
ribosomal protein L34 |
| chr15_+_59374198 | 1.35 |
ENSMUST00000079703.3
ENSMUST00000168722.1 |
Nsmce2
|
non-SMC element 2 homolog (MMS21, S. cerevisiae) |
| chr8_+_70234613 | 1.31 |
ENSMUST00000145078.1
|
Sugp2
|
SURP and G patch domain containing 2 |
| chr8_-_69184177 | 1.28 |
ENSMUST00000185176.1
|
Lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr18_-_34624562 | 1.28 |
ENSMUST00000003876.3
ENSMUST00000115766.1 ENSMUST00000097626.3 ENSMUST00000115765.1 |
Brd8
|
bromodomain containing 8 |
| chr19_-_5366285 | 1.27 |
ENSMUST00000170010.1
|
Banf1
|
barrier to autointegration factor 1 |
| chr12_-_55821157 | 1.27 |
ENSMUST00000110687.1
ENSMUST00000085385.5 |
Ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 |
| chr8_-_69791170 | 1.26 |
ENSMUST00000131784.1
|
Zfp866
|
zinc finger protein 866 |
| chr17_-_57011271 | 1.25 |
ENSMUST00000002733.6
|
Gtf2f1
|
general transcription factor IIF, polypeptide 1 |
| chr13_+_95325195 | 1.24 |
ENSMUST00000045909.7
|
Zbed3
|
zinc finger, BED domain containing 3 |
| chr3_-_69004503 | 1.22 |
ENSMUST00000107812.1
|
Ift80
|
intraflagellar transport 80 |
| chr7_-_139978709 | 1.20 |
ENSMUST00000121412.1
|
6430531B16Rik
|
RIKEN cDNA 6430531B16 gene |
| chr2_-_120731503 | 1.19 |
ENSMUST00000110701.1
ENSMUST00000110700.1 |
Cdan1
|
congenital dyserythropoietic anemia, type I (human) |
| chr16_+_14163275 | 1.19 |
ENSMUST00000023359.6
ENSMUST00000117958.1 |
Nde1
|
nuclear distribution gene E homolog 1 (A nidulans) |
| chr5_-_122372230 | 1.19 |
ENSMUST00000031419.5
|
Fam216a
|
family with sequence similarity 216, member A |
| chr3_-_95995999 | 1.18 |
ENSMUST00000015889.3
|
Plekho1
|
pleckstrin homology domain containing, family O member 1 |
| chr3_-_95995835 | 1.18 |
ENSMUST00000143485.1
|
Plekho1
|
pleckstrin homology domain containing, family O member 1 |
| chrX_-_57393020 | 1.16 |
ENSMUST00000143310.1
ENSMUST00000098470.2 ENSMUST00000114726.1 |
Rbmx
|
RNA binding motif protein, X chromosome |
| chr7_+_102065485 | 1.16 |
ENSMUST00000106950.1
ENSMUST00000146450.1 |
Trpc2
|
transient receptor potential cation channel, subfamily C, member 2 |
| chr17_+_35841491 | 1.16 |
ENSMUST00000082337.6
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr5_-_149053038 | 1.15 |
ENSMUST00000085546.6
|
Hmgb1
|
high mobility group box 1 |
| chr4_-_115133977 | 1.14 |
ENSMUST00000051400.7
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr14_+_31001383 | 1.14 |
ENSMUST00000168584.1
|
Glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr15_-_55072139 | 1.13 |
ENSMUST00000041733.7
|
Taf2
|
TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr8_-_107439585 | 1.12 |
ENSMUST00000077208.4
|
Rps26-ps1
|
ribosomal protein S26, pseudogene 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mybl2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0002884 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) negative regulation of hypersensitivity(GO:0002884) |
| 2.4 | 9.5 | GO:0045575 | basophil activation(GO:0045575) |
| 2.3 | 6.9 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 1.5 | 5.8 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 1.4 | 7.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 1.4 | 5.7 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 1.3 | 3.9 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 1.3 | 11.7 | GO:0002432 | granuloma formation(GO:0002432) |
| 1.3 | 3.8 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 1.2 | 3.5 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 1.1 | 5.6 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 1.1 | 3.3 | GO:0071395 | sesquiterpenoid metabolic process(GO:0006714) response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 1.1 | 5.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 1.0 | 7.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.9 | 20.5 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.9 | 4.6 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.9 | 5.6 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.8 | 3.3 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.8 | 3.2 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.8 | 2.4 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.8 | 2.3 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.7 | 3.7 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.7 | 3.7 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.7 | 7.8 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.7 | 12.0 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.7 | 2.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.6 | 2.6 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.6 | 11.7 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.6 | 1.8 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.6 | 2.4 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.6 | 2.9 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) histone H3-S10 phosphorylation(GO:0043987) |
| 0.6 | 15.7 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.6 | 2.8 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.5 | 1.6 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.5 | 5.4 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.5 | 2.7 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.5 | 2.7 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.5 | 1.6 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.5 | 2.7 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.5 | 13.8 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.5 | 3.4 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.5 | 2.4 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.4 | 16.3 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.4 | 1.3 | GO:0070676 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.4 | 4.7 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.4 | 3.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.4 | 8.6 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.4 | 6.5 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.4 | 2.8 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.4 | 12.1 | GO:0007099 | centriole replication(GO:0007099) |
| 0.4 | 1.9 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.4 | 3.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.4 | 2.5 | GO:0044838 | cell quiescence(GO:0044838) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.4 | 1.4 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.3 | 4.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.3 | 2.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.3 | 6.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 2.0 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.3 | 1.0 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.3 | 0.9 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.3 | 4.3 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.3 | 4.8 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.3 | 1.4 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.3 | 2.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.3 | 9.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.3 | 1.0 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.3 | 1.8 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 | 4.0 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.2 | 1.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.2 | 2.2 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.2 | 1.7 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.2 | 1.0 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.2 | 0.9 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 0.6 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.2 | 2.9 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.2 | 3.0 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.2 | 0.8 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.2 | 2.3 | GO:0048757 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.2 | 2.3 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 3.1 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.2 | 11.9 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.2 | 2.6 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 0.9 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.2 | 2.7 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.2 | 1.8 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 0.5 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 1.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 1.6 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.2 | 6.4 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.2 | 0.5 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 8.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 5.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.4 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.1 | 0.6 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.1 | 1.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 1.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.9 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.1 | 0.9 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 4.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.8 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 2.4 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 2.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.5 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 0.8 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.4 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 0.5 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.4 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 3.1 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 1.9 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.8 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.3 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.1 | 1.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.6 | GO:0048549 | endosome localization(GO:0032439) positive regulation of pinocytosis(GO:0048549) Golgi to lysosome transport(GO:0090160) |
| 0.1 | 5.2 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.7 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.1 | 0.8 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 1.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.0 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.1 | 0.3 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.5 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.1 | 1.2 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 0.9 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 1.6 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 1.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 9.3 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 1.7 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 4.4 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 1.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 4.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.9 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 1.8 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 4.5 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.6 | GO:0002070 | epithelial cell maturation(GO:0002070) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.7 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.8 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 1.4 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 1.0 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.6 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 2.5 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.0 | 0.3 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 1.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.1 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.8 | GO:0006997 | nucleus organization(GO:0006997) |
| 0.0 | 0.5 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 2.3 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 2.5 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.5 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 0.0 | 0.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 18.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 1.6 | 15.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 1.3 | 7.9 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.1 | 3.2 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 1.1 | 11.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 1.0 | 8.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.8 | 3.9 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.8 | 3.1 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.7 | 5.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.7 | 2.8 | GO:0032585 | multivesicular body membrane(GO:0032585) multivesicular body, internal vesicle(GO:0097487) |
| 0.7 | 4.8 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.7 | 8.6 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.6 | 3.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.6 | 5.4 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.6 | 6.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.6 | 4.6 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.4 | 3.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.4 | 3.8 | GO:0032591 | dendritic spine membrane(GO:0032591) dendritic spine neck(GO:0044326) |
| 0.3 | 3.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.3 | 0.9 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.3 | 1.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 1.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.3 | 1.4 | GO:1990923 | PET complex(GO:1990923) |
| 0.3 | 2.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.3 | 1.6 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 5.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 5.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 2.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 1.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.2 | 12.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 5.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 4.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 6.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 0.5 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.2 | 0.9 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.2 | 0.8 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.1 | 1.0 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 7.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 2.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 5.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.8 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.9 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 2.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 2.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.9 | GO:0097526 | spliceosomal tri-snRNP complex(GO:0097526) |
| 0.1 | 4.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.8 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 2.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.9 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 2.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 5.6 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.8 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 0.9 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.3 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.1 | 0.3 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 3.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 3.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.9 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 1.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 12.5 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 1.1 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 0.6 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 2.7 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 0.9 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 0.7 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 5.6 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 11.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.0 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 4.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 1.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 7.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 1.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 2.9 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 11.7 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 3.2 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.9 | GO:0016459 | myosin complex(GO:0016459) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 20.5 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 2.6 | 7.8 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 2.3 | 6.9 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 1.3 | 4.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 1.2 | 3.5 | GO:0000401 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 1.1 | 3.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 1.1 | 3.3 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 1.1 | 4.3 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.9 | 4.6 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.9 | 6.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.8 | 3.9 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.8 | 2.4 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.6 | 1.9 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.6 | 4.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.5 | 4.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.5 | 2.6 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.5 | 5.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.5 | 2.9 | GO:0035184 | nucleosomal histone binding(GO:0031493) histone threonine kinase activity(GO:0035184) |
| 0.5 | 1.4 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.4 | 1.8 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.4 | 0.9 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.4 | 5.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.4 | 1.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 3.8 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.3 | 0.9 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.3 | 0.9 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.3 | 1.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.3 | 7.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.3 | 3.7 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 17.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 2.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 1.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 5.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 5.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 6.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 0.8 | GO:0002135 | CTP binding(GO:0002135) |
| 0.2 | 2.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 10.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.2 | 0.9 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 1.3 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.2 | 0.5 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.2 | 3.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.2 | 0.5 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.2 | 1.4 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.2 | 0.8 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 0.9 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 1.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 8.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 7.3 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 6.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 11.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 3.7 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 2.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.7 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 2.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 6.6 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 7.7 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 2.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.6 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 5.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 2.4 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.1 | 17.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 0.9 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.9 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 2.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 1.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 1.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 4.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 6.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 2.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.9 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 5.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 1.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.5 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.8 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.0 | 6.9 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 8.4 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.8 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 1.3 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 1.4 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 3.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 2.1 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 4.5 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 2.0 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 6.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.5 | 24.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.5 | 7.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.4 | 15.2 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 9.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 3.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 5.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 3.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 2.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 2.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 9.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 2.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.7 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.5 | 19.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.5 | 7.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.4 | 11.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.4 | 7.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.4 | 40.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.3 | 2.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.3 | 9.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.3 | 7.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 10.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 2.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 3.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 3.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.2 | 3.1 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.2 | 2.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 4.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 6.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 1.9 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 7.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 5.6 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 2.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 3.0 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 2.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 7.6 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 4.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 12.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 2.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 5.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.9 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.0 | 5.4 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 10.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 1.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 2.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 3.0 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |