Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Mef2b
Z-value: 0.79
Transcription factors associated with Mef2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mef2b
|
ENSMUSG00000079033.3 | myocyte enhancer factor 2B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mef2b | mm10_v2_chr8_+_70152754_70152781 | 0.59 | 1.6e-04 | Click! |
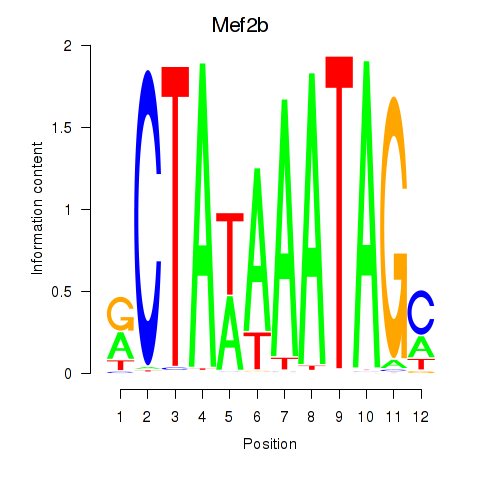
Activity profile of Mef2b motif
Sorted Z-values of Mef2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_46039202 | 5.68 |
ENSMUST00000156200.1
|
Tmod1
|
tropomodulin 1 |
| chr14_-_63417125 | 3.99 |
ENSMUST00000014597.3
|
Blk
|
B lymphoid kinase |
| chr6_+_41458923 | 3.90 |
ENSMUST00000031910.7
|
Prss1
|
protease, serine, 1 (trypsin 1) |
| chr1_-_66935333 | 3.73 |
ENSMUST00000120415.1
ENSMUST00000119429.1 |
Myl1
|
myosin, light polypeptide 1 |
| chr3_-_123034943 | 3.39 |
ENSMUST00000029761.7
|
Myoz2
|
myozenin 2 |
| chr2_+_148798785 | 3.32 |
ENSMUST00000028931.3
ENSMUST00000109947.1 |
Cst8
|
cystatin 8 (cystatin-related epididymal spermatogenic) |
| chr15_-_89425856 | 3.29 |
ENSMUST00000109313.2
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr7_-_103843154 | 3.20 |
ENSMUST00000063957.4
|
Hbb-bh1
|
hemoglobin Z, beta-like embryonic chain |
| chr6_+_87778084 | 3.13 |
ENSMUST00000032133.3
|
Gp9
|
glycoprotein 9 (platelet) |
| chr7_+_142441808 | 2.94 |
ENSMUST00000105971.1
ENSMUST00000145287.1 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr7_+_142442330 | 2.85 |
ENSMUST00000149529.1
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr17_+_71019548 | 2.80 |
ENSMUST00000073211.5
ENSMUST00000179759.1 |
Myom1
|
myomesin 1 |
| chr10_-_88605017 | 2.75 |
ENSMUST00000119185.1
ENSMUST00000121629.1 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr11_-_69948145 | 2.70 |
ENSMUST00000179298.1
ENSMUST00000018710.6 ENSMUST00000135437.1 ENSMUST00000141837.2 ENSMUST00000142500.1 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr17_+_71019503 | 2.65 |
ENSMUST00000024847.7
|
Myom1
|
myomesin 1 |
| chr14_-_20656488 | 2.51 |
ENSMUST00000090469.6
|
Myoz1
|
myozenin 1 |
| chr11_+_67798269 | 2.43 |
ENSMUST00000168612.1
ENSMUST00000040574.4 |
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr7_+_99594605 | 2.30 |
ENSMUST00000162290.1
|
Arrb1
|
arrestin, beta 1 |
| chrX_-_93832106 | 2.29 |
ENSMUST00000045748.6
|
Pdk3
|
pyruvate dehydrogenase kinase, isoenzyme 3 |
| chr8_-_46211284 | 2.22 |
ENSMUST00000034049.4
|
Slc25a4
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 |
| chrX_+_68761839 | 2.20 |
ENSMUST00000069731.5
|
Fmr1nb
|
fragile X mental retardation 1 neighbor |
| chr7_-_128206346 | 2.18 |
ENSMUST00000033049.7
|
Cox6a2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chrX_+_157699113 | 2.17 |
ENSMUST00000112521.1
|
Smpx
|
small muscle protein, X-linked |
| chr7_+_45335256 | 2.17 |
ENSMUST00000085351.4
|
Hrc
|
histidine rich calcium binding protein |
| chr4_+_49059256 | 2.08 |
ENSMUST00000076670.2
|
E130309F12Rik
|
RIKEN cDNA E130309F12 gene |
| chr5_+_122100951 | 1.99 |
ENSMUST00000014080.6
ENSMUST00000111750.1 ENSMUST00000139213.1 ENSMUST00000111751.1 ENSMUST00000155612.1 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr15_+_79891631 | 1.98 |
ENSMUST00000177350.1
ENSMUST00000177483.1 |
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr15_-_58324161 | 1.96 |
ENSMUST00000022985.1
|
Klhl38
|
kelch-like 38 |
| chr8_+_15057646 | 1.94 |
ENSMUST00000033842.3
|
Myom2
|
myomesin 2 |
| chr15_-_89425795 | 1.84 |
ENSMUST00000168376.1
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr3_+_102086471 | 1.83 |
ENSMUST00000165540.2
ENSMUST00000164123.1 |
Casq2
|
calsequestrin 2 |
| chr11_-_94976327 | 1.83 |
ENSMUST00000103162.1
ENSMUST00000166320.1 |
Sgca
|
sarcoglycan, alpha (dystrophin-associated glycoprotein) |
| chr7_-_14123042 | 1.78 |
ENSMUST00000098809.2
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr3_+_102086415 | 1.75 |
ENSMUST00000029454.5
|
Casq2
|
calsequestrin 2 |
| chr7_-_100514800 | 1.67 |
ENSMUST00000054923.7
|
Dnajb13
|
DnaJ (Hsp40) related, subfamily B, member 13 |
| chr17_+_5799491 | 1.61 |
ENSMUST00000181484.1
|
3300005D01Rik
|
RIKEN cDNA 3300005D01 gene |
| chr11_+_75656103 | 1.61 |
ENSMUST00000136935.1
|
Myo1c
|
myosin IC |
| chr1_-_172219715 | 1.59 |
ENSMUST00000170700.1
ENSMUST00000003554.4 |
Casq1
|
calsequestrin 1 |
| chr5_-_148392810 | 1.58 |
ENSMUST00000138257.1
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr9_+_121777607 | 1.57 |
ENSMUST00000098272.2
|
Klhl40
|
kelch-like 40 |
| chr18_+_60963517 | 1.57 |
ENSMUST00000115295.2
ENSMUST00000039904.6 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr1_-_45503282 | 1.53 |
ENSMUST00000086430.4
|
Col5a2
|
collagen, type V, alpha 2 |
| chrX_+_159303266 | 1.52 |
ENSMUST00000112491.1
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chrX_+_68761890 | 1.49 |
ENSMUST00000071848.6
|
Fmr1nb
|
fragile X mental retardation 1 neighbor |
| chr7_+_81862674 | 1.37 |
ENSMUST00000119543.1
|
Tm6sf1
|
transmembrane 6 superfamily member 1 |
| chrX_+_68761875 | 1.34 |
ENSMUST00000114647.1
|
Fmr1nb
|
fragile X mental retardation 1 neighbor |
| chr4_+_134315112 | 1.34 |
ENSMUST00000105875.1
ENSMUST00000030638.6 |
Trim63
|
tripartite motif-containing 63 |
| chr17_+_5799616 | 1.32 |
ENSMUST00000181392.1
|
3300005D01Rik
|
RIKEN cDNA 3300005D01 gene |
| chr4_+_156215920 | 1.31 |
ENSMUST00000105572.1
|
2310042D19Rik
|
RIKEN cDNA 2310042D19 gene |
| chr3_+_40800013 | 1.30 |
ENSMUST00000026858.5
ENSMUST00000170825.1 |
Plk4
|
polo-like kinase 4 |
| chr2_+_120463566 | 1.30 |
ENSMUST00000028749.7
ENSMUST00000110721.1 |
Capn3
|
calpain 3 |
| chr14_-_37110087 | 1.27 |
ENSMUST00000179488.1
|
2610528A11Rik
|
RIKEN cDNA 2610528A11 gene |
| chr6_-_126939524 | 1.27 |
ENSMUST00000144954.1
ENSMUST00000112221.1 ENSMUST00000112220.1 |
Rad51ap1
|
RAD51 associated protein 1 |
| chr7_-_30823766 | 1.24 |
ENSMUST00000053156.3
|
Ffar2
|
free fatty acid receptor 2 |
| chr10_+_119992962 | 1.23 |
ENSMUST00000154238.1
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr3_-_98814434 | 1.23 |
ENSMUST00000029463.6
|
Hsd3b6
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 6 |
| chrX_+_164438039 | 1.22 |
ENSMUST00000033755.5
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr5_+_25759987 | 1.19 |
ENSMUST00000128727.1
ENSMUST00000088244.4 |
Actr3b
|
ARP3 actin-related protein 3B |
| chr11_+_104577281 | 1.18 |
ENSMUST00000106956.3
|
Myl4
|
myosin, light polypeptide 4 |
| chr6_-_136941887 | 1.18 |
ENSMUST00000111891.1
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr4_+_5724304 | 1.14 |
ENSMUST00000108380.1
|
Fam110b
|
family with sequence similarity 110, member B |
| chr18_+_50030977 | 1.11 |
ENSMUST00000145726.1
ENSMUST00000128377.1 |
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chrX_+_157698910 | 1.10 |
ENSMUST00000136141.1
|
Smpx
|
small muscle protein, X-linked |
| chrX_-_95478107 | 1.10 |
ENSMUST00000033549.2
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr14_+_26894557 | 1.07 |
ENSMUST00000090337.4
ENSMUST00000165929.2 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr4_-_73950834 | 1.05 |
ENSMUST00000095023.1
ENSMUST00000030101.3 |
2310002L09Rik
|
RIKEN cDNA 2310002L09 gene |
| chr15_+_94629148 | 1.04 |
ENSMUST00000080141.4
|
Tmem117
|
transmembrane protein 117 |
| chr11_+_104576965 | 1.04 |
ENSMUST00000106957.1
|
Myl4
|
myosin, light polypeptide 4 |
| chr3_+_146852359 | 1.00 |
ENSMUST00000038090.5
ENSMUST00000170055.1 |
Ttll7
|
tubulin tyrosine ligase-like family, member 7 |
| chr7_+_30121915 | 0.99 |
ENSMUST00000098596.3
ENSMUST00000153792.1 |
Zfp382
|
zinc finger protein 382 |
| chr7_-_102100227 | 0.94 |
ENSMUST00000106937.1
|
Art5
|
ADP-ribosyltransferase 5 |
| chr13_-_113663670 | 0.93 |
ENSMUST00000054650.4
|
Hspb3
|
heat shock protein 3 |
| chr3_+_32817520 | 0.92 |
ENSMUST00000072312.5
ENSMUST00000108228.1 |
Usp13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr1_+_75375271 | 0.89 |
ENSMUST00000087122.5
|
Speg
|
SPEG complex locus |
| chr17_-_14694223 | 0.89 |
ENSMUST00000170872.1
|
Thbs2
|
thrombospondin 2 |
| chr7_+_91090728 | 0.87 |
ENSMUST00000074273.3
|
Dlg2
|
discs, large homolog 2 (Drosophila) |
| chr6_-_136941694 | 0.86 |
ENSMUST00000032344.5
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr15_+_91673175 | 0.86 |
ENSMUST00000060642.6
|
Lrrk2
|
leucine-rich repeat kinase 2 |
| chr6_+_48589445 | 0.85 |
ENSMUST00000064744.3
|
Gm5111
|
predicted gene 5111 |
| chr1_-_4360256 | 0.85 |
ENSMUST00000027032.4
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chrX_+_142825698 | 0.85 |
ENSMUST00000112888.1
|
Tmem164
|
transmembrane protein 164 |
| chr15_+_80133114 | 0.84 |
ENSMUST00000023050.7
|
Tab1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr13_+_20090538 | 0.82 |
ENSMUST00000072519.5
|
Elmo1
|
engulfment and cell motility 1 |
| chr8_+_31089471 | 0.82 |
ENSMUST00000036631.7
ENSMUST00000170204.1 |
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr2_+_24186469 | 0.80 |
ENSMUST00000057567.2
|
Il1f9
|
interleukin 1 family, member 9 |
| chr7_+_91090697 | 0.80 |
ENSMUST00000107196.2
|
Dlg2
|
discs, large homolog 2 (Drosophila) |
| chr18_+_21072329 | 0.80 |
ENSMUST00000082235.4
|
Mep1b
|
meprin 1 beta |
| chr19_-_36919606 | 0.80 |
ENSMUST00000057337.7
|
Fgfbp3
|
fibroblast growth factor binding protein 3 |
| chr14_-_20668269 | 0.78 |
ENSMUST00000057090.5
ENSMUST00000117386.1 |
Synpo2l
|
synaptopodin 2-like |
| chr17_+_34039437 | 0.78 |
ENSMUST00000131134.1
ENSMUST00000087497.4 ENSMUST00000114255.1 ENSMUST00000114252.1 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr5_+_105519388 | 0.77 |
ENSMUST00000067924.6
ENSMUST00000150981.1 |
Lrrc8c
|
leucine rich repeat containing 8 family, member C |
| chr5_+_66745835 | 0.73 |
ENSMUST00000101164.4
ENSMUST00000118242.1 ENSMUST00000119854.1 ENSMUST00000117601.1 |
Limch1
|
LIM and calponin homology domains 1 |
| chr2_+_91096744 | 0.72 |
ENSMUST00000132741.2
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr13_+_20090500 | 0.71 |
ENSMUST00000165249.2
|
Elmo1
|
engulfment and cell motility 1 |
| chrX_-_47892502 | 0.71 |
ENSMUST00000077569.4
ENSMUST00000101616.2 ENSMUST00000088973.4 |
Smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr3_-_56183678 | 0.71 |
ENSMUST00000029374.6
|
Nbea
|
neurobeachin |
| chr11_-_99155067 | 0.71 |
ENSMUST00000103134.3
|
Ccr7
|
chemokine (C-C motif) receptor 7 |
| chr16_+_11313812 | 0.70 |
ENSMUST00000023140.5
|
Tnfrsf17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr10_+_33863935 | 0.69 |
ENSMUST00000092597.3
|
Sult3a1
|
sulfotransferase family 3A, member 1 |
| chr14_-_21714570 | 0.65 |
ENSMUST00000073870.5
|
Dupd1
|
dual specificity phosphatase and pro isomerase domain containing 1 |
| chr6_+_112273758 | 0.65 |
ENSMUST00000032376.5
|
Lmcd1
|
LIM and cysteine-rich domains 1 |
| chr18_+_34409415 | 0.64 |
ENSMUST00000166156.1
ENSMUST00000014647.7 |
Pkd2l2
|
polycystic kidney disease 2-like 2 |
| chr11_+_70657196 | 0.63 |
ENSMUST00000157027.1
ENSMUST00000072841.5 ENSMUST00000108548.1 ENSMUST00000126241.1 |
Eno3
|
enolase 3, beta muscle |
| chr2_-_26516620 | 0.63 |
ENSMUST00000132820.1
|
Notch1
|
notch 1 |
| chr16_-_23127702 | 0.62 |
ENSMUST00000115338.1
ENSMUST00000115337.1 ENSMUST00000023598.8 |
Rfc4
|
replication factor C (activator 1) 4 |
| chr3_-_150073620 | 0.62 |
ENSMUST00000057740.5
|
Rpsa-ps10
|
ribosomal protein SA, pseudogene 10 |
| chr7_-_142899985 | 0.62 |
ENSMUST00000000219.3
|
Th
|
tyrosine hydroxylase |
| chr17_+_47726834 | 0.60 |
ENSMUST00000024782.5
ENSMUST00000144955.1 |
Pgc
|
progastricsin (pepsinogen C) |
| chrX_+_75096039 | 0.60 |
ENSMUST00000131155.1
ENSMUST00000132000.1 |
Dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr18_+_76059458 | 0.60 |
ENSMUST00000167921.1
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr10_+_21593151 | 0.59 |
ENSMUST00000057341.4
|
1700020N01Rik
|
RIKEN cDNA 1700020N01 gene |
| chr4_+_62663620 | 0.59 |
ENSMUST00000126338.1
|
Rgs3
|
regulator of G-protein signaling 3 |
| chr13_+_5861489 | 0.58 |
ENSMUST00000000080.6
|
Klf6
|
Kruppel-like factor 6 |
| chr15_-_50889691 | 0.58 |
ENSMUST00000165201.2
ENSMUST00000184458.1 |
Trps1
|
trichorhinophalangeal syndrome I (human) |
| chr1_+_187608755 | 0.55 |
ENSMUST00000127489.1
|
Esrrg
|
estrogen-related receptor gamma |
| chr19_+_8723478 | 0.55 |
ENSMUST00000180819.1
ENSMUST00000181422.1 |
Snhg1
|
small nucleolar RNA host gene (non-protein coding) 1 |
| chr3_-_96814518 | 0.54 |
ENSMUST00000047702.7
|
Cd160
|
CD160 antigen |
| chr15_+_101266839 | 0.52 |
ENSMUST00000023779.6
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr4_+_119108795 | 0.51 |
ENSMUST00000134105.1
ENSMUST00000144329.1 |
Slc2a1
|
solute carrier family 2 (facilitated glucose transporter), member 1 |
| chr11_+_70657687 | 0.51 |
ENSMUST00000134087.1
ENSMUST00000170716.1 |
Eno3
|
enolase 3, beta muscle |
| chr1_-_138175238 | 0.50 |
ENSMUST00000182536.1
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr11_-_79504078 | 0.46 |
ENSMUST00000164465.2
|
Omg
|
oligodendrocyte myelin glycoprotein |
| chr8_-_34965631 | 0.46 |
ENSMUST00000033929.4
|
Tnks
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr2_+_43748802 | 0.45 |
ENSMUST00000112824.1
ENSMUST00000055776.7 |
Arhgap15
|
Rho GTPase activating protein 15 |
| chr2_+_172440556 | 0.45 |
ENSMUST00000029005.3
|
Rtfdc1
|
replication termination factor 2 domain containing 1 |
| chr5_-_3893907 | 0.45 |
ENSMUST00000117463.1
ENSMUST00000044746.4 |
Mterf
|
mitochondrial transcription termination factor |
| chrX_+_75095854 | 0.44 |
ENSMUST00000033776.8
|
Dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr1_-_138175283 | 0.44 |
ENSMUST00000182755.1
ENSMUST00000183262.1 ENSMUST00000027645.7 ENSMUST00000112036.2 ENSMUST00000182283.1 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr5_+_114896936 | 0.44 |
ENSMUST00000031542.9
ENSMUST00000146072.1 ENSMUST00000150361.1 |
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr8_+_81342556 | 0.42 |
ENSMUST00000172167.1
ENSMUST00000169116.1 ENSMUST00000109852.2 ENSMUST00000172031.1 |
Inpp4b
|
inositol polyphosphate-4-phosphatase, type II |
| chr9_+_99629496 | 0.42 |
ENSMUST00000131095.1
ENSMUST00000078367.5 ENSMUST00000112885.2 |
Dzip1l
|
DAZ interacting protein 1-like |
| chrY_-_1286563 | 0.42 |
ENSMUST00000091190.5
|
Ddx3y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked |
| chr11_-_83649349 | 0.42 |
ENSMUST00000001008.5
|
Ccl3
|
chemokine (C-C motif) ligand 3 |
| chr4_-_131967824 | 0.41 |
ENSMUST00000146443.1
ENSMUST00000135579.1 |
Epb4.1
|
erythrocyte protein band 4.1 |
| chr1_-_138175126 | 0.41 |
ENSMUST00000183301.1
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr10_-_128549125 | 0.41 |
ENSMUST00000177163.1
ENSMUST00000176683.1 ENSMUST00000176010.1 |
Rpl41
|
ribosomal protein L41 |
| chr7_+_128003911 | 0.40 |
ENSMUST00000106248.1
|
Trim72
|
tripartite motif-containing 72 |
| chr3_+_96833218 | 0.40 |
ENSMUST00000128789.1
|
Pdzk1
|
PDZ domain containing 1 |
| chr10_+_33083476 | 0.38 |
ENSMUST00000095762.4
|
Trdn
|
triadin |
| chr3_-_123236134 | 0.38 |
ENSMUST00000106427.1
ENSMUST00000106426.1 ENSMUST00000051443.5 |
Synpo2
|
synaptopodin 2 |
| chr10_-_69212996 | 0.37 |
ENSMUST00000170048.1
|
A930033H14Rik
|
RIKEN cDNA A930033H14 gene |
| chr15_-_81843699 | 0.36 |
ENSMUST00000092020.2
|
Gm8444
|
predicted gene 8444 |
| chr10_-_128549102 | 0.35 |
ENSMUST00000176906.1
|
Rpl41
|
ribosomal protein L41 |
| chr10_-_129902726 | 0.35 |
ENSMUST00000071557.1
|
Olfr815
|
olfactory receptor 815 |
| chr5_+_8660059 | 0.34 |
ENSMUST00000047753.4
|
Abcb1a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1A |
| chr5_+_129096740 | 0.33 |
ENSMUST00000056617.7
ENSMUST00000156437.1 |
Gpr133
|
G protein-coupled receptor 133 |
| chr16_+_57353093 | 0.31 |
ENSMUST00000159816.1
|
Filip1l
|
filamin A interacting protein 1-like |
| chr9_+_69397933 | 0.31 |
ENSMUST00000117610.1
ENSMUST00000145538.1 ENSMUST00000117246.1 |
Narg2
|
NMDA receptor-regulated gene 2 |
| chr15_-_80083374 | 0.31 |
ENSMUST00000081650.7
|
Rpl3
|
ribosomal protein L3 |
| chr3_+_14533817 | 0.31 |
ENSMUST00000169079.1
ENSMUST00000091325.3 |
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr6_+_69600066 | 0.30 |
ENSMUST00000114209.1
|
Gm11143
|
predicted gene 11143 |
| chr1_+_78816909 | 0.30 |
ENSMUST00000057262.6
|
Kcne4
|
potassium voltage-gated channel, Isk-related subfamily, gene 4 |
| chr9_+_99629823 | 0.29 |
ENSMUST00000112886.2
|
Dzip1l
|
DAZ interacting protein 1-like |
| chr5_-_111761697 | 0.28 |
ENSMUST00000129146.1
ENSMUST00000137398.1 ENSMUST00000129065.1 |
E130006D01Rik
|
RIKEN cDNA E130006D01 gene |
| chr3_+_14533788 | 0.27 |
ENSMUST00000108370.2
|
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr11_+_102268732 | 0.27 |
ENSMUST00000036467.4
|
Asb16
|
ankyrin repeat and SOCS box-containing 16 |
| chr17_+_29032664 | 0.26 |
ENSMUST00000130216.1
|
Srsf3
|
serine/arginine-rich splicing factor 3 |
| chr9_+_57072024 | 0.26 |
ENSMUST00000169879.1
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
| chr11_+_75655873 | 0.26 |
ENSMUST00000108431.2
|
Myo1c
|
myosin IC |
| chr5_-_16731074 | 0.25 |
ENSMUST00000073014.5
|
Gm8991
|
predicted pseudogene 8991 |
| chr16_+_95702044 | 0.25 |
ENSMUST00000023612.8
|
Ets2
|
E26 avian leukemia oncogene 2, 3' domain |
| chr2_-_79908389 | 0.25 |
ENSMUST00000090756.4
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr18_+_67464849 | 0.24 |
ENSMUST00000025411.7
|
Slmo1
|
slowmo homolog 1 (Drosophila) |
| chr9_-_108649349 | 0.24 |
ENSMUST00000013338.8
|
Arih2
|
ariadne homolog 2 (Drosophila) |
| chr3_-_154597045 | 0.24 |
ENSMUST00000052774.1
ENSMUST00000170461.1 ENSMUST00000122976.1 |
Tyw3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr1_-_88509886 | 0.23 |
ENSMUST00000067625.7
|
Glrp1
|
glutamine repeat protein 1 |
| chr19_-_46969474 | 0.22 |
ENSMUST00000086961.7
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr2_-_79908428 | 0.22 |
ENSMUST00000102652.3
ENSMUST00000102651.3 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr12_-_111672290 | 0.21 |
ENSMUST00000001304.7
|
Ckb
|
creatine kinase, brain |
| chr1_-_173942445 | 0.21 |
ENSMUST00000042228.8
ENSMUST00000081216.5 ENSMUST00000129829.1 ENSMUST00000123708.1 ENSMUST00000111210.2 |
Ifi203
Mndal
|
interferon activated gene 203 myeloid nuclear differentiation antigen like |
| chr5_-_35575046 | 0.21 |
ENSMUST00000030980.7
|
Trmt44
|
tRNA methyltransferase 44 |
| chr3_+_68584154 | 0.20 |
ENSMUST00000182997.1
|
Schip1
|
schwannomin interacting protein 1 |
| chr17_+_91236787 | 0.20 |
ENSMUST00000057074.8
|
Gm6741
|
predicted gene 6741 |
| chr4_+_119108711 | 0.20 |
ENSMUST00000030398.3
|
Slc2a1
|
solute carrier family 2 (facilitated glucose transporter), member 1 |
| chr7_-_12818837 | 0.20 |
ENSMUST00000121215.1
ENSMUST00000108546.1 ENSMUST00000072222.7 |
Zfp329
|
zinc finger protein 329 |
| chr11_+_54304005 | 0.20 |
ENSMUST00000000145.5
ENSMUST00000138515.1 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr11_+_54304180 | 0.19 |
ENSMUST00000108904.3
ENSMUST00000108905.3 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr9_-_64022027 | 0.19 |
ENSMUST00000179458.1
|
Smad6
|
SMAD family member 6 |
| chr18_+_80206887 | 0.19 |
ENSMUST00000127234.1
|
Gm16286
|
predicted gene 16286 |
| chr1_-_191183244 | 0.19 |
ENSMUST00000027941.8
|
Atf3
|
activating transcription factor 3 |
| chr17_+_15499888 | 0.19 |
ENSMUST00000159197.1
ENSMUST00000162505.1 ENSMUST00000014911.5 ENSMUST00000147081.2 ENSMUST00000118001.1 ENSMUST00000143924.1 ENSMUST00000119879.2 ENSMUST00000155051.1 ENSMUST00000117593.1 |
Tbp
|
TATA box binding protein |
| chr11_+_77462325 | 0.18 |
ENSMUST00000102493.1
|
Coro6
|
coronin 6 |
| chr13_-_23368969 | 0.18 |
ENSMUST00000152557.1
|
Zfp322a
|
zinc finger protein 322A |
| chr3_-_20155069 | 0.17 |
ENSMUST00000184552.1
ENSMUST00000178328.1 |
Gyg
|
glycogenin |
| chr2_-_80128834 | 0.17 |
ENSMUST00000102654.4
ENSMUST00000102655.3 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr2_+_118663235 | 0.17 |
ENSMUST00000099557.3
|
Pak6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr2_-_126876209 | 0.17 |
ENSMUST00000103224.3
|
Trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr8_+_19682268 | 0.16 |
ENSMUST00000153710.1
ENSMUST00000127799.1 |
Gm6483
|
predicted gene 6483 |
| chr2_-_126876117 | 0.16 |
ENSMUST00000028843.5
|
Trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr3_+_14533867 | 0.16 |
ENSMUST00000163660.1
|
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr3_-_65958236 | 0.15 |
ENSMUST00000029416.7
|
Ccnl1
|
cyclin L1 |
| chr7_+_101896817 | 0.15 |
ENSMUST00000143835.1
|
Anapc15
|
anaphase prompoting complex C subunit 15 |
| chr14_-_50897456 | 0.14 |
ENSMUST00000170855.1
|
A930018M24Rik
|
RIKEN cDNA A930018M24 gene |
| chr12_-_111980751 | 0.14 |
ENSMUST00000170525.1
|
BC048943
|
cDNA sequence BC048943 |
| chr6_+_126939957 | 0.14 |
ENSMUST00000032497.3
|
D6Wsu163e
|
DNA segment, Chr 6, Wayne State University 163, expressed |
| chr6_+_137410721 | 0.13 |
ENSMUST00000167002.1
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr11_+_95723587 | 0.13 |
ENSMUST00000100534.2
|
B130006D01Rik
|
RIKEN cDNA B130006D01 gene |
| chr3_+_137341067 | 0.13 |
ENSMUST00000122064.1
|
Emcn
|
endomucin |
| chr19_-_40402267 | 0.12 |
ENSMUST00000099467.3
ENSMUST00000099466.3 ENSMUST00000165212.1 ENSMUST00000165469.1 |
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr1_-_74600564 | 0.11 |
ENSMUST00000127938.1
ENSMUST00000154874.1 |
Rnf25
|
ring finger protein 25 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mef2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 15.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.7 | 2.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.6 | 2.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.6 | 1.7 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.5 | 1.6 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.5 | 3.6 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.4 | 1.3 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.4 | 1.3 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.4 | 1.2 | GO:0002879 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.4 | 3.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.4 | 2.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.4 | 1.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 | 1.7 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.3 | 1.6 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.3 | 1.9 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.3 | 0.9 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.3 | 0.9 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.3 | 0.8 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.3 | 1.3 | GO:2000473 | immunoglobulin biosynthetic process(GO:0002378) regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.3 | 2.0 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.2 | 0.7 | GO:2000547 | lymphocyte migration into lymphoid organs(GO:0097021) positive regulation of thymocyte migration(GO:2000412) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.2 | 1.0 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) telomerase RNA stabilization(GO:0090669) |
| 0.2 | 0.8 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.2 | 1.3 | GO:0098535 | positive regulation of centriole replication(GO:0046601) de novo centriole assembly(GO:0098535) |
| 0.2 | 2.2 | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) |
| 0.2 | 2.8 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.2 | 0.6 | GO:1902263 | inhibition of neuroepithelial cell differentiation(GO:0002085) atrioventricular node development(GO:0003162) coronary vein morphogenesis(GO:0003169) negative regulation of auditory receptor cell differentiation(GO:0045608) regulation of cell adhesion involved in heart morphogenesis(GO:0061344) positive regulation of ephrin receptor signaling pathway(GO:1901189) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.2 | 2.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 0.8 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 0.5 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 0.7 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 0.7 | GO:0042908 | xenobiotic transport(GO:0042908) dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 1.6 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 0.6 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.1 | 0.6 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 5.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.6 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 | 1.7 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.1 | 0.8 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 1.6 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 2.7 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.1 | 0.6 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 1.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.3 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 0.3 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.3 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 | 1.3 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 2.2 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 0.8 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 0.6 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 4.7 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 5.3 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.1 | 0.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 4.0 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 0.4 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 6.9 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 0.2 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.6 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.4 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.6 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 4.8 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 0.4 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.4 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.3 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.7 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 1.5 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.2 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 3.2 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.1 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 1.1 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.5 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.3 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.8 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.2 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.3 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.5 | 1.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.5 | 3.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.5 | 1.9 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.4 | 3.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.4 | 1.6 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.4 | 10.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.3 | 5.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 0.9 | GO:0044753 | amphisome(GO:0044753) |
| 0.3 | 1.0 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.2 | 5.7 | GO:0036379 | myofilament(GO:0036379) |
| 0.2 | 1.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 1.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 1.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 4.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 0.7 | GO:0090537 | CERF complex(GO:0090537) |
| 0.2 | 2.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 2.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 2.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.6 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 1.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.6 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 3.8 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.9 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.1 | 0.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 7.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 3.7 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.9 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 7.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 4.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.4 | GO:0043292 | contractile fiber(GO:0043292) |
| 0.0 | 1.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.7 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.9 | GO:0051373 | FATZ binding(GO:0051373) |
| 1.0 | 5.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.8 | 3.2 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.8 | 2.3 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.7 | 3.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.6 | 5.8 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.6 | 2.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.5 | 10.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.5 | 2.0 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.4 | 5.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 0.9 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.3 | 0.8 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.3 | 2.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 1.8 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.2 | 1.0 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 0.7 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 0.2 | 0.9 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.2 | 0.6 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.2 | 1.6 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.2 | 2.6 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 2.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 0.9 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 1.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 1.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 2.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 4.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.6 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 2.2 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 5.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 1.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.3 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 0.4 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 0.2 | GO:0030617 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) |
| 0.1 | 0.8 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.2 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.1 | 4.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.6 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 2.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 3.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 1.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 2.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 2.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 6.9 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 4.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 3.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 5.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 2.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 3.0 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 2.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 6.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.5 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 21.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 3.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 3.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 5.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 2.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 3.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 4.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 2.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 1.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 1.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 2.2 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 1.5 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 3.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.5 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.4 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 2.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |