Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
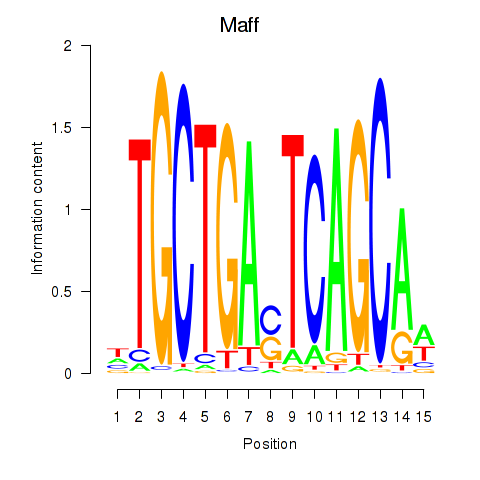
Results for Maff
Z-value: 0.89
Transcription factors associated with Maff
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Maff
|
ENSMUSG00000042622.7 | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Maff | mm10_v2_chr15_+_79347534_79347556 | 0.61 | 7.0e-05 | Click! |
Activity profile of Maff motif
Sorted Z-values of Maff motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_88205674 | 7.45 |
ENSMUST00000119972.2
|
Dnajb3
|
DnaJ (Hsp40) homolog, subfamily B, member 3 |
| chr19_+_58728887 | 7.13 |
ENSMUST00000048644.5
|
Pnliprp1
|
pancreatic lipase related protein 1 |
| chr6_-_40999479 | 6.07 |
ENSMUST00000166306.1
|
Gm2663
|
predicted gene 2663 |
| chr6_+_40964760 | 5.67 |
ENSMUST00000076638.5
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr5_-_67815852 | 5.20 |
ENSMUST00000141443.1
|
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
| chr5_+_106964319 | 4.99 |
ENSMUST00000031221.5
ENSMUST00000117196.2 ENSMUST00000076467.6 |
Cdc7
|
cell division cycle 7 (S. cerevisiae) |
| chr7_+_35449035 | 4.94 |
ENSMUST00000118969.1
ENSMUST00000118383.1 |
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr14_+_118787894 | 4.38 |
ENSMUST00000047761.6
ENSMUST00000071546.7 |
Cldn10
|
claudin 10 |
| chr7_+_35449154 | 3.46 |
ENSMUST00000032703.9
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chrX_+_93675088 | 3.29 |
ENSMUST00000045898.3
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr6_-_136401830 | 3.21 |
ENSMUST00000058713.7
|
E330021D16Rik
|
RIKEN cDNA E330021D16 gene |
| chr6_-_78378851 | 3.12 |
ENSMUST00000089667.1
ENSMUST00000167492.1 |
Reg3d
|
regenerating islet-derived 3 delta |
| chr2_-_127831817 | 3.04 |
ENSMUST00000028858.7
|
Bub1
|
budding uninhibited by benzimidazoles 1 homolog (S. cerevisiae) |
| chr5_+_140505550 | 2.68 |
ENSMUST00000043050.8
ENSMUST00000124142.1 |
Chst12
|
carbohydrate sulfotransferase 12 |
| chr15_-_78572754 | 2.44 |
ENSMUST00000043214.6
|
Rac2
|
RAS-related C3 botulinum substrate 2 |
| chr7_-_110862944 | 2.35 |
ENSMUST00000033050.3
|
Lyve1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr10_-_88605017 | 2.17 |
ENSMUST00000119185.1
ENSMUST00000121629.1 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr7_+_46861203 | 2.15 |
ENSMUST00000014545.4
|
Ldhc
|
lactate dehydrogenase C |
| chr18_+_60963517 | 1.95 |
ENSMUST00000115295.2
ENSMUST00000039904.6 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr2_-_69206146 | 1.86 |
ENSMUST00000127243.1
ENSMUST00000149643.1 ENSMUST00000167875.2 ENSMUST00000005365.8 |
Spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr2_-_69206133 | 1.85 |
ENSMUST00000112320.1
|
Spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr7_+_30493622 | 1.68 |
ENSMUST00000058280.6
ENSMUST00000133318.1 ENSMUST00000142575.1 ENSMUST00000131040.1 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr3_+_109573907 | 1.66 |
ENSMUST00000106576.2
|
Vav3
|
vav 3 oncogene |
| chr11_-_109473598 | 1.52 |
ENSMUST00000070152.5
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr11_+_82101836 | 1.50 |
ENSMUST00000000194.3
|
Ccl12
|
chemokine (C-C motif) ligand 12 |
| chr7_+_84528964 | 1.47 |
ENSMUST00000180387.1
|
Gm2115
|
predicted gene 2115 |
| chr17_-_67950908 | 1.40 |
ENSMUST00000164647.1
|
Arhgap28
|
Rho GTPase activating protein 28 |
| chr11_+_119229092 | 1.38 |
ENSMUST00000053440.7
|
Ccdc40
|
coiled-coil domain containing 40 |
| chr2_-_20943413 | 1.33 |
ENSMUST00000140230.1
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr15_-_25413752 | 1.31 |
ENSMUST00000058845.7
|
Basp1
|
brain abundant, membrane attached signal protein 1 |
| chr1_+_75382114 | 1.25 |
ENSMUST00000113590.1
ENSMUST00000148515.1 |
Speg
|
SPEG complex locus |
| chr18_+_46850034 | 1.16 |
ENSMUST00000025358.2
|
4833403I15Rik
|
RIKEN cDNA 4833403I15 gene |
| chr7_-_64925560 | 1.16 |
ENSMUST00000149851.1
|
Fam189a1
|
family with sequence similarity 189, member A1 |
| chr9_-_111690313 | 1.16 |
ENSMUST00000035083.7
|
Stac
|
src homology three (SH3) and cysteine rich domain |
| chr12_+_16894894 | 1.06 |
ENSMUST00000020904.6
|
Rock2
|
Rho-associated coiled-coil containing protein kinase 2 |
| chr7_-_100964371 | 0.93 |
ENSMUST00000060174.4
|
P2ry6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr8_-_12672135 | 0.93 |
ENSMUST00000000776.8
|
Tubgcp3
|
tubulin, gamma complex associated protein 3 |
| chr5_-_135251209 | 0.92 |
ENSMUST00000062572.2
|
Fzd9
|
frizzled homolog 9 (Drosophila) |
| chr5_+_110839973 | 0.90 |
ENSMUST00000066160.1
|
Chek2
|
checkpoint kinase 2 |
| chr5_+_103754560 | 0.84 |
ENSMUST00000153165.1
ENSMUST00000031256.5 |
Aff1
|
AF4/FMR2 family, member 1 |
| chr17_+_86963279 | 0.82 |
ENSMUST00000139344.1
|
Rhoq
|
ras homolog gene family, member Q |
| chr1_-_161979636 | 0.75 |
ENSMUST00000162676.1
|
4930558K02Rik
|
RIKEN cDNA 4930558K02 gene |
| chr14_+_66784523 | 0.74 |
ENSMUST00000071522.2
|
Gm10032
|
predicted gene 10032 |
| chr18_+_65582390 | 0.74 |
ENSMUST00000169679.1
ENSMUST00000183326.1 |
Zfp532
|
zinc finger protein 532 |
| chr3_-_59262825 | 0.69 |
ENSMUST00000050360.7
|
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr18_-_63387124 | 0.67 |
ENSMUST00000047480.6
ENSMUST00000046860.5 |
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chr19_+_56722372 | 0.63 |
ENSMUST00000038949.4
|
Adrb1
|
adrenergic receptor, beta 1 |
| chr15_+_99972780 | 0.61 |
ENSMUST00000100206.2
|
Larp4
|
La ribonucleoprotein domain family, member 4 |
| chr6_+_120773633 | 0.56 |
ENSMUST00000112682.2
|
Slc25a18
|
solute carrier family 25 (mitochondrial carrier), member 18 |
| chr7_+_16944645 | 0.56 |
ENSMUST00000094807.5
|
Pnmal2
|
PNMA-like 2 |
| chr17_-_25570678 | 0.52 |
ENSMUST00000025003.3
ENSMUST00000173447.1 |
Sox8
|
SRY-box containing gene 8 |
| chr4_+_94556546 | 0.50 |
ENSMUST00000094969.1
|
Gm10306
|
predicted gene 10306 |
| chr14_+_20929416 | 0.50 |
ENSMUST00000022369.7
|
Vcl
|
vinculin |
| chr5_-_35525691 | 0.49 |
ENSMUST00000038676.5
|
Cpz
|
carboxypeptidase Z |
| chr18_+_33464163 | 0.43 |
ENSMUST00000097634.3
|
Gm10549
|
predicted gene 10549 |
| chr5_+_7179299 | 0.40 |
ENSMUST00000179460.1
|
Tubb4b-ps1
|
tubulin, beta 4B class IVB, pseudogene 1 |
| chr7_+_44052290 | 0.38 |
ENSMUST00000079859.5
|
Klk1b27
|
kallikrein 1-related peptidase b27 |
| chr4_-_143299463 | 0.37 |
ENSMUST00000119654.1
|
Pdpn
|
podoplanin |
| chr11_+_75651504 | 0.35 |
ENSMUST00000069057.6
|
Myo1c
|
myosin IC |
| chr13_-_54749849 | 0.30 |
ENSMUST00000135343.1
|
Gprin1
|
G protein-regulated inducer of neurite outgrowth 1 |
| chr1_-_75232093 | 0.26 |
ENSMUST00000180101.1
|
A630095N17Rik
|
RIKEN cDNA A630095N17 gene |
| chr6_+_71293765 | 0.25 |
ENSMUST00000114185.2
|
Gm1070
|
predicted gene 1070 |
| chr17_+_27839974 | 0.25 |
ENSMUST00000071006.7
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr1_+_136017967 | 0.24 |
ENSMUST00000063719.8
ENSMUST00000118832.1 |
Tmem9
|
transmembrane protein 9 |
| chr4_-_94556737 | 0.23 |
ENSMUST00000030313.8
|
Caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr3_+_127791374 | 0.23 |
ENSMUST00000171621.1
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr2_+_121866918 | 0.21 |
ENSMUST00000078752.3
ENSMUST00000110586.3 |
Casc4
|
cancer susceptibility candidate 4 |
| chr3_+_28805436 | 0.19 |
ENSMUST00000043867.5
|
Rpl22l1
|
ribosomal protein L22 like 1 |
| chr11_-_118342500 | 0.19 |
ENSMUST00000103024.3
|
BC100451
|
cDNA sequence BC100451 |
| chr5_+_8056527 | 0.18 |
ENSMUST00000148633.1
|
Sri
|
sorcin |
| chr18_-_33463747 | 0.17 |
ENSMUST00000171533.1
|
Nrep
|
neuronal regeneration related protein |
| chr7_+_46861407 | 0.17 |
ENSMUST00000126004.1
|
Ldhc
|
lactate dehydrogenase C |
| chr7_+_30314810 | 0.16 |
ENSMUST00000054594.8
ENSMUST00000177078.1 ENSMUST00000176504.1 ENSMUST00000176304.1 |
Syne4
|
spectrin repeat containing, nuclear envelope family member 4 |
| chr4_-_136886187 | 0.12 |
ENSMUST00000046384.8
|
C1qb
|
complement component 1, q subcomponent, beta polypeptide |
| chr2_+_121867083 | 0.12 |
ENSMUST00000089912.5
ENSMUST00000089915.3 |
Casc4
|
cancer susceptibility candidate 4 |
| chr3_-_108154871 | 0.12 |
ENSMUST00000062028.1
|
Gpr61
|
G protein-coupled receptor 61 |
| chr8_+_25532125 | 0.09 |
ENSMUST00000167764.1
|
Fgfr1
|
fibroblast growth factor receptor 1 |
| chr13_+_3924686 | 0.09 |
ENSMUST00000021639.6
|
Tubal3
|
tubulin, alpha-like 3 |
| chr11_+_83709015 | 0.08 |
ENSMUST00000001009.7
|
Wfdc18
|
WAP four-disulfide core domain 18 |
| chr18_-_33464007 | 0.08 |
ENSMUST00000168890.1
|
Nrep
|
neuronal regeneration related protein |
| chr12_+_104263117 | 0.08 |
ENSMUST00000109958.2
|
Serpina3i
|
serine (or cysteine) peptidase inhibitor, clade A, member 3I |
| chr10_-_121626316 | 0.07 |
ENSMUST00000039810.7
|
Xpot
|
exportin, tRNA (nuclear export receptor for tRNAs) |
| chr7_+_46861253 | 0.07 |
ENSMUST00000148565.1
|
Ldhc
|
lactate dehydrogenase C |
| chr5_-_35525567 | 0.06 |
ENSMUST00000132959.1
|
Cpz
|
carboxypeptidase Z |
| chr6_+_83795022 | 0.06 |
ENSMUST00000113851.1
|
Nagk
|
N-acetylglucosamine kinase |
| chr16_-_22439719 | 0.06 |
ENSMUST00000079601.6
|
Etv5
|
ets variant gene 5 |
| chr9_+_65460926 | 0.06 |
ENSMUST00000034955.6
|
Spg21
|
spastic paraplegia 21 homolog (human) |
| chr11_+_76945719 | 0.05 |
ENSMUST00000125145.1
|
Blmh
|
bleomycin hydrolase |
| chr9_-_57765845 | 0.04 |
ENSMUST00000065330.6
|
Clk3
|
CDC-like kinase 3 |
| chr13_-_54749627 | 0.02 |
ENSMUST00000099506.1
|
Gprin1
|
G protein-regulated inducer of neurite outgrowth 1 |
| chr6_+_83794974 | 0.02 |
ENSMUST00000037376.7
|
Nagk
|
N-acetylglucosamine kinase |
| chr18_+_32377176 | 0.01 |
ENSMUST00000091967.5
ENSMUST00000025239.7 |
Bin1
|
bridging integrator 1 |
| chr17_+_26138661 | 0.01 |
ENSMUST00000074370.3
ENSMUST00000118904.2 ENSMUST00000163421.1 |
Axin1
|
axin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Maff
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.4 | GO:0015811 | L-cystine transport(GO:0015811) |
| 1.0 | 3.0 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.9 | 2.7 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.8 | 5.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.7 | 5.2 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.6 | 1.7 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.5 | 2.4 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.4 | 1.5 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.3 | 1.4 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.3 | 0.9 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.3 | 2.4 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.3 | 0.9 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 0.3 | 1.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 1.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 | 1.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 0.6 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.2 | 0.5 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.2 | 0.7 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 | 2.0 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 2.4 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 2.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.9 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 0.4 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.9 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 1.3 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 0.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 1.7 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 3.3 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 3.7 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.5 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:1903463 | regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 0.0 | 0.2 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.0 | 0.1 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.7 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 1.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 1.3 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.7 | 3.7 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.5 | 2.0 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.1 | 0.9 | GO:0005827 | equatorial microtubule organizing center(GO:0000923) polar microtubule(GO:0005827) |
| 0.1 | 0.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 5.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 2.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 8.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 1.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 7.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 1.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 5.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.4 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 3.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) nuclear membrane part(GO:0044453) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.4 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.9 | 2.7 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.7 | 3.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.4 | 7.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.4 | 1.5 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.3 | 2.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 0.9 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.2 | 2.0 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 5.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 0.6 | GO:0031694 | beta-adrenergic receptor activity(GO:0004939) alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 2.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 2.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.9 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 12.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.6 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 1.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 10.6 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 4.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 2.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 5.0 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 2.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 0.9 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 5.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 3.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 0.9 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 2.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 6.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.7 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |