Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Hoxb7
Z-value: 1.96
Transcription factors associated with Hoxb7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb7
|
ENSMUSG00000038721.8 | homeobox B7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb7 | mm10_v2_chr11_+_96286623_96286653 | -0.01 | 9.7e-01 | Click! |
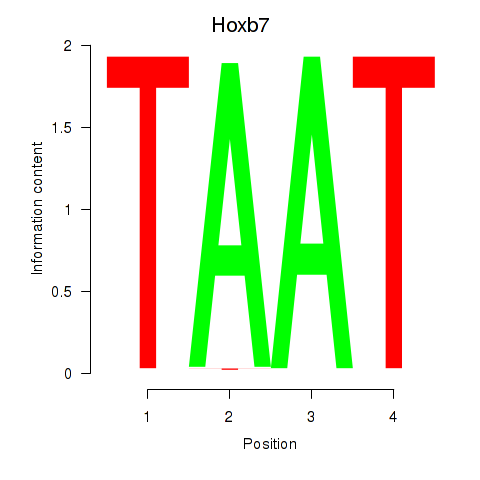
Activity profile of Hoxb7 motif
Sorted Z-values of Hoxb7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_75270073 | 5.06 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr5_-_62765618 | 4.62 |
ENSMUST00000159470.1
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr11_-_30198232 | 4.15 |
ENSMUST00000102838.3
|
Sptbn1
|
spectrin beta, non-erythrocytic 1 |
| chr4_+_108719649 | 4.10 |
ENSMUST00000178992.1
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr18_+_20247340 | 4.05 |
ENSMUST00000054128.6
|
Dsg1c
|
desmoglein 1 gamma |
| chr8_-_36953139 | 4.04 |
ENSMUST00000179501.1
|
Dlc1
|
deleted in liver cancer 1 |
| chr1_-_150466165 | 3.92 |
ENSMUST00000162367.1
ENSMUST00000161611.1 ENSMUST00000161320.1 ENSMUST00000159035.1 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr3_-_113532288 | 3.61 |
ENSMUST00000132353.1
|
Amy2a1
|
amylase 2a1 |
| chr8_-_8639363 | 3.54 |
ENSMUST00000152698.1
|
Efnb2
|
ephrin B2 |
| chr2_-_62483637 | 3.54 |
ENSMUST00000136686.1
ENSMUST00000102733.3 |
Gcg
|
glucagon |
| chr4_+_102589687 | 3.44 |
ENSMUST00000097949.4
ENSMUST00000106901.1 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr19_+_26749726 | 3.26 |
ENSMUST00000175842.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr19_+_58670358 | 3.23 |
ENSMUST00000057270.7
|
Pnlip
|
pancreatic lipase |
| chr19_+_26605106 | 3.22 |
ENSMUST00000025862.7
ENSMUST00000176030.1 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_+_179961110 | 3.11 |
ENSMUST00000076687.5
ENSMUST00000097450.3 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr1_+_177445660 | 3.05 |
ENSMUST00000077225.6
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr8_+_45507768 | 3.01 |
ENSMUST00000067065.7
ENSMUST00000098788.3 ENSMUST00000067107.7 ENSMUST00000171337.2 ENSMUST00000138049.1 ENSMUST00000141039.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr8_+_45627709 | 2.98 |
ENSMUST00000134321.1
ENSMUST00000135336.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr19_+_26750939 | 2.95 |
ENSMUST00000175953.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr6_+_15196949 | 2.92 |
ENSMUST00000151301.1
ENSMUST00000131414.1 ENSMUST00000140557.1 ENSMUST00000115469.1 |
Foxp2
|
forkhead box P2 |
| chr3_+_5218516 | 2.83 |
ENSMUST00000175866.1
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr3_+_41563356 | 2.81 |
ENSMUST00000163764.1
|
Phf17
|
PHD finger protein 17 |
| chr11_+_67200137 | 2.81 |
ENSMUST00000129018.1
|
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr8_-_67974567 | 2.69 |
ENSMUST00000098696.3
ENSMUST00000038959.9 ENSMUST00000093469.4 |
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr2_-_45112890 | 2.68 |
ENSMUST00000076836.6
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr12_+_59095653 | 2.59 |
ENSMUST00000021384.4
|
Mia2
|
melanoma inhibitory activity 2 |
| chr15_+_25773985 | 2.55 |
ENSMUST00000125667.1
|
Myo10
|
myosin X |
| chr1_-_150465563 | 2.54 |
ENSMUST00000164600.1
ENSMUST00000111902.2 ENSMUST00000111901.2 ENSMUST00000006171.9 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr5_+_42067960 | 2.46 |
ENSMUST00000087332.4
|
Gm16223
|
predicted gene 16223 |
| chr6_+_29859686 | 2.45 |
ENSMUST00000134438.1
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr6_+_30541582 | 2.45 |
ENSMUST00000096066.4
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr14_-_36935560 | 2.44 |
ENSMUST00000183038.1
|
Ccser2
|
coiled-coil serine rich 2 |
| chr2_+_68104671 | 2.42 |
ENSMUST00000042456.3
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr3_+_60081861 | 2.41 |
ENSMUST00000029326.5
|
Sucnr1
|
succinate receptor 1 |
| chr6_+_17491216 | 2.38 |
ENSMUST00000080469.5
|
Met
|
met proto-oncogene |
| chr1_+_187997835 | 2.36 |
ENSMUST00000110938.1
|
Esrrg
|
estrogen-related receptor gamma |
| chr6_+_29859662 | 2.34 |
ENSMUST00000128927.2
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr1_-_30863256 | 2.33 |
ENSMUST00000088310.3
|
Phf3
|
PHD finger protein 3 |
| chr5_+_89027959 | 2.31 |
ENSMUST00000130041.1
|
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4 |
| chr1_-_139608282 | 2.25 |
ENSMUST00000170441.2
|
Cfhr3
|
complement factor H-related 3 |
| chr10_-_20724696 | 2.24 |
ENSMUST00000170265.1
|
Pde7b
|
phosphodiesterase 7B |
| chr1_-_165934900 | 2.24 |
ENSMUST00000069609.5
ENSMUST00000111427.2 ENSMUST00000111426.4 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr11_+_94044111 | 2.24 |
ENSMUST00000132079.1
|
Spag9
|
sperm associated antigen 9 |
| chr16_+_43247278 | 2.20 |
ENSMUST00000114691.1
ENSMUST00000079441.6 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr6_+_29859374 | 2.20 |
ENSMUST00000115238.3
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr6_-_144209471 | 2.17 |
ENSMUST00000038815.7
|
Sox5
|
SRY-box containing gene 5 |
| chr9_+_120929216 | 2.14 |
ENSMUST00000130466.1
|
Ctnnb1
|
catenin (cadherin associated protein), beta 1 |
| chr6_+_97991776 | 2.13 |
ENSMUST00000043628.6
|
Mitf
|
microphthalmia-associated transcription factor |
| chr2_+_101624696 | 2.12 |
ENSMUST00000044031.3
|
Rag2
|
recombination activating gene 2 |
| chr11_+_67277124 | 2.12 |
ENSMUST00000019625.5
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr19_+_39287074 | 2.09 |
ENSMUST00000003137.8
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr10_-_109009055 | 2.08 |
ENSMUST00000156979.1
|
Syt1
|
synaptotagmin I |
| chr6_-_144209558 | 2.06 |
ENSMUST00000111749.1
ENSMUST00000170367.2 |
Sox5
|
SRY-box containing gene 5 |
| chr6_-_144209448 | 2.03 |
ENSMUST00000077160.5
|
Sox5
|
SRY-box containing gene 5 |
| chr5_-_62766153 | 2.02 |
ENSMUST00000076623.4
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr5_-_66080971 | 2.01 |
ENSMUST00000127275.1
ENSMUST00000113724.1 |
Rbm47
|
RNA binding motif protein 47 |
| chr8_-_107065632 | 2.00 |
ENSMUST00000034393.5
|
Tmed6
|
transmembrane emp24 protein transport domain containing 6 |
| chr14_-_36919314 | 1.99 |
ENSMUST00000182797.1
|
Ccser2
|
coiled-coil serine rich 2 |
| chr5_-_123141067 | 1.98 |
ENSMUST00000162697.1
ENSMUST00000160321.1 ENSMUST00000159637.1 |
AI480526
|
expressed sequence AI480526 |
| chr13_+_49582745 | 1.97 |
ENSMUST00000065494.7
|
Omd
|
osteomodulin |
| chr3_-_79145875 | 1.96 |
ENSMUST00000118340.1
|
Rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr19_+_26748268 | 1.94 |
ENSMUST00000175791.1
ENSMUST00000176698.1 ENSMUST00000177252.1 ENSMUST00000176475.1 ENSMUST00000112637.2 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr13_-_101692624 | 1.94 |
ENSMUST00000035532.6
|
Pik3r1
|
phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha) |
| chr3_-_20275659 | 1.93 |
ENSMUST00000011607.5
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr8_-_5105232 | 1.92 |
ENSMUST00000023835.1
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr14_+_17981633 | 1.92 |
ENSMUST00000022304.8
|
Thrb
|
thyroid hormone receptor beta |
| chr6_+_15185456 | 1.91 |
ENSMUST00000115472.1
ENSMUST00000115474.1 ENSMUST00000031545.7 ENSMUST00000137628.1 |
Foxp2
|
forkhead box P2 |
| chr18_+_20376723 | 1.90 |
ENSMUST00000076737.6
|
Dsg1b
|
desmoglein 1 beta |
| chr14_+_101840501 | 1.90 |
ENSMUST00000159026.1
|
Lmo7
|
LIM domain only 7 |
| chr12_+_38783455 | 1.88 |
ENSMUST00000161980.1
ENSMUST00000160701.1 |
Etv1
|
ets variant gene 1 |
| chr3_-_146770218 | 1.87 |
ENSMUST00000106137.1
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr14_+_28511344 | 1.84 |
ENSMUST00000112272.1
|
Wnt5a
|
wingless-related MMTV integration site 5A |
| chr3_+_115080965 | 1.84 |
ENSMUST00000051309.8
|
Olfm3
|
olfactomedin 3 |
| chr8_-_67818284 | 1.83 |
ENSMUST00000120071.1
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr8_-_41016749 | 1.82 |
ENSMUST00000117735.1
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr10_+_127420334 | 1.81 |
ENSMUST00000171434.1
|
R3hdm2
|
R3H domain containing 2 |
| chr13_+_49544443 | 1.80 |
ENSMUST00000177948.1
ENSMUST00000021820.6 |
Aspn
|
asporin |
| chr7_-_70366735 | 1.79 |
ENSMUST00000089565.5
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr1_-_87394721 | 1.78 |
ENSMUST00000113212.3
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr11_-_99374895 | 1.77 |
ENSMUST00000006963.2
|
Krt28
|
keratin 28 |
| chr9_+_53301571 | 1.76 |
ENSMUST00000051014.1
|
Exph5
|
exophilin 5 |
| chr1_-_162898665 | 1.74 |
ENSMUST00000111510.1
ENSMUST00000045902.6 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr6_-_138079916 | 1.73 |
ENSMUST00000171804.1
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr18_+_20310738 | 1.73 |
ENSMUST00000077146.3
|
Dsg1a
|
desmoglein 1 alpha |
| chr12_+_38780284 | 1.72 |
ENSMUST00000162563.1
ENSMUST00000161164.1 ENSMUST00000160996.1 |
Etv1
|
ets variant gene 1 |
| chr6_+_15185203 | 1.71 |
ENSMUST00000154448.1
|
Foxp2
|
forkhead box P2 |
| chr6_-_41035501 | 1.70 |
ENSMUST00000031931.5
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr3_-_116712696 | 1.70 |
ENSMUST00000169530.1
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr15_+_25742314 | 1.68 |
ENSMUST00000135981.1
|
Myo10
|
myosin X |
| chr3_-_116712644 | 1.67 |
ENSMUST00000029569.2
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr3_-_146770603 | 1.67 |
ENSMUST00000106138.1
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr3_+_5218546 | 1.67 |
ENSMUST00000026284.6
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr14_+_79515618 | 1.67 |
ENSMUST00000110835.1
|
Elf1
|
E74-like factor 1 |
| chr3_-_59220150 | 1.65 |
ENSMUST00000170388.1
|
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr18_-_66022580 | 1.65 |
ENSMUST00000143990.1
|
Lman1
|
lectin, mannose-binding, 1 |
| chr10_+_42583787 | 1.64 |
ENSMUST00000105497.1
ENSMUST00000144806.1 |
Ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr4_+_99295900 | 1.63 |
ENSMUST00000094955.1
|
Gm12689
|
predicted gene 12689 |
| chr5_+_89028035 | 1.61 |
ENSMUST00000113216.2
ENSMUST00000134303.1 |
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4 |
| chrX_+_166344692 | 1.60 |
ENSMUST00000112223.1
ENSMUST00000112224.1 ENSMUST00000112229.2 ENSMUST00000112228.1 ENSMUST00000112227.2 ENSMUST00000112226.2 |
Gpm6b
|
glycoprotein m6b |
| chrM_+_11734 | 1.59 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr2_-_77703252 | 1.59 |
ENSMUST00000171063.1
|
Zfp385b
|
zinc finger protein 385B |
| chr2_+_76650264 | 1.58 |
ENSMUST00000099986.2
|
Dfnb59
|
deafness, autosomal recessive 59 (human) |
| chr2_-_132111440 | 1.58 |
ENSMUST00000128899.1
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr17_-_90088343 | 1.58 |
ENSMUST00000173917.1
|
Nrxn1
|
neurexin I |
| chr19_+_39007019 | 1.58 |
ENSMUST00000025966.4
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr2_+_102658640 | 1.57 |
ENSMUST00000080210.3
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr3_-_30140407 | 1.57 |
ENSMUST00000108271.3
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr1_-_136960427 | 1.57 |
ENSMUST00000027649.7
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr11_+_94044331 | 1.56 |
ENSMUST00000024979.8
|
Spag9
|
sperm associated antigen 9 |
| chr11_+_94044241 | 1.55 |
ENSMUST00000103168.3
|
Spag9
|
sperm associated antigen 9 |
| chr12_-_56613270 | 1.55 |
ENSMUST00000072631.5
|
Nkx2-9
|
NK2 homeobox 9 |
| chr2_+_23069210 | 1.55 |
ENSMUST00000155602.1
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr3_+_66985700 | 1.54 |
ENSMUST00000046542.6
ENSMUST00000162693.1 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr10_+_127898515 | 1.54 |
ENSMUST00000047134.7
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr2_-_77519565 | 1.54 |
ENSMUST00000111830.2
|
Zfp385b
|
zinc finger protein 385B |
| chr2_-_35061431 | 1.54 |
ENSMUST00000028233.3
|
Hc
|
hemolytic complement |
| chr11_+_96024540 | 1.53 |
ENSMUST00000103157.3
|
Gip
|
gastric inhibitory polypeptide |
| chr7_-_119459266 | 1.53 |
ENSMUST00000033255.5
|
Gp2
|
glycoprotein 2 (zymogen granule membrane) |
| chr1_-_162898484 | 1.53 |
ENSMUST00000143123.1
|
Fmo2
|
flavin containing monooxygenase 2 |
| chr11_+_94044194 | 1.52 |
ENSMUST00000092777.4
ENSMUST00000075695.6 |
Spag9
|
sperm associated antigen 9 |
| chr9_-_101198999 | 1.52 |
ENSMUST00000066773.7
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr4_-_76344227 | 1.51 |
ENSMUST00000050757.9
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D |
| chr11_-_62392605 | 1.51 |
ENSMUST00000151498.2
ENSMUST00000159069.1 |
Ncor1
|
nuclear receptor co-repressor 1 |
| chr12_+_38783503 | 1.50 |
ENSMUST00000159334.1
|
Etv1
|
ets variant gene 1 |
| chr1_-_24612700 | 1.50 |
ENSMUST00000088336.1
|
Gm10222
|
predicted gene 10222 |
| chr18_-_39487096 | 1.50 |
ENSMUST00000097592.2
ENSMUST00000115571.1 |
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr12_-_34528844 | 1.49 |
ENSMUST00000110819.2
|
Hdac9
|
histone deacetylase 9 |
| chrM_+_9870 | 1.49 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr2_+_3704787 | 1.49 |
ENSMUST00000115054.2
|
Fam107b
|
family with sequence similarity 107, member B |
| chr16_-_64786321 | 1.49 |
ENSMUST00000052588.4
|
Zfp654
|
zinc finger protein 654 |
| chr9_+_75775355 | 1.49 |
ENSMUST00000012281.7
|
Bmp5
|
bone morphogenetic protein 5 |
| chr2_-_7081207 | 1.48 |
ENSMUST00000114923.2
ENSMUST00000182706.1 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr7_-_73541738 | 1.47 |
ENSMUST00000169922.2
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr10_+_97479470 | 1.47 |
ENSMUST00000105287.3
|
Dcn
|
decorin |
| chr5_-_53707532 | 1.47 |
ENSMUST00000031093.3
|
Cckar
|
cholecystokinin A receptor |
| chr3_-_113258837 | 1.45 |
ENSMUST00000098673.3
|
Amy2a5
|
amylase 2a5 |
| chr2_+_20737306 | 1.44 |
ENSMUST00000114606.1
ENSMUST00000114608.1 |
Etl4
|
enhancer trap locus 4 |
| chr8_-_60954726 | 1.43 |
ENSMUST00000110302.1
|
Clcn3
|
chloride channel 3 |
| chr2_-_18048347 | 1.43 |
ENSMUST00000066885.5
|
Skida1
|
SKI/DACH domain containing 1 |
| chr18_+_51117754 | 1.42 |
ENSMUST00000116639.2
|
Prr16
|
proline rich 16 |
| chr16_+_56204313 | 1.42 |
ENSMUST00000160116.1
ENSMUST00000069936.7 |
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr7_+_112742025 | 1.41 |
ENSMUST00000164363.1
|
Tead1
|
TEA domain family member 1 |
| chr4_-_41517326 | 1.41 |
ENSMUST00000030152.6
ENSMUST00000095126.4 |
1110017D15Rik
|
RIKEN cDNA 1110017D15 gene |
| chr9_-_29963112 | 1.41 |
ENSMUST00000075069.4
|
Ntm
|
neurotrimin |
| chr10_-_20725023 | 1.40 |
ENSMUST00000020165.7
|
Pde7b
|
phosphodiesterase 7B |
| chr9_+_13765970 | 1.40 |
ENSMUST00000152532.1
|
Mtmr2
|
myotubularin related protein 2 |
| chr1_+_187997821 | 1.39 |
ENSMUST00000027906.6
|
Esrrg
|
estrogen-related receptor gamma |
| chr1_-_166002613 | 1.38 |
ENSMUST00000177358.1
ENSMUST00000160908.1 ENSMUST00000027850.8 ENSMUST00000160260.2 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr6_+_34598500 | 1.37 |
ENSMUST00000079391.3
ENSMUST00000142512.1 |
Cald1
|
caldesmon 1 |
| chr4_-_141846277 | 1.37 |
ENSMUST00000105781.1
|
Ctrc
|
chymotrypsin C (caldecrin) |
| chr3_+_66985647 | 1.37 |
ENSMUST00000162362.1
ENSMUST00000065074.7 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr1_-_4360256 | 1.36 |
ENSMUST00000027032.4
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chrX_+_107149580 | 1.36 |
ENSMUST00000137107.1
ENSMUST00000067249.2 |
A630033H20Rik
|
RIKEN cDNA A630033H20 gene |
| chr13_+_93308006 | 1.36 |
ENSMUST00000079086.6
|
Homer1
|
homer homolog 1 (Drosophila) |
| chr10_+_34483400 | 1.35 |
ENSMUST00000019913.7
ENSMUST00000170771.1 |
Frk
|
fyn-related kinase |
| chr2_-_7396192 | 1.35 |
ENSMUST00000137733.2
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr4_+_102421518 | 1.34 |
ENSMUST00000106904.2
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr9_-_62070606 | 1.34 |
ENSMUST00000034785.7
|
Glce
|
glucuronyl C5-epimerase |
| chr4_+_103143052 | 1.34 |
ENSMUST00000106855.1
|
Mier1
|
mesoderm induction early response 1 homolog (Xenopus laevis |
| chr5_-_146220901 | 1.34 |
ENSMUST00000169407.2
ENSMUST00000161331.1 ENSMUST00000159074.2 ENSMUST00000067837.3 |
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr5_-_87569023 | 1.34 |
ENSMUST00000113314.2
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr9_-_80465429 | 1.33 |
ENSMUST00000085289.5
ENSMUST00000185068.1 ENSMUST00000113250.3 |
Impg1
|
interphotoreceptor matrix proteoglycan 1 |
| chr5_-_123140135 | 1.33 |
ENSMUST00000160099.1
|
AI480526
|
expressed sequence AI480526 |
| chr3_+_66985680 | 1.33 |
ENSMUST00000065047.6
|
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr10_+_26822560 | 1.33 |
ENSMUST00000135866.1
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chrY_+_90843934 | 1.33 |
ENSMUST00000178550.1
|
Gm21742
|
predicted gene, 21742 |
| chrX_-_160994665 | 1.33 |
ENSMUST00000087104.4
|
Cdkl5
|
cyclin-dependent kinase-like 5 |
| chr2_+_3770673 | 1.32 |
ENSMUST00000177037.1
|
Fam107b
|
family with sequence similarity 107, member B |
| chr6_+_141629499 | 1.30 |
ENSMUST00000042812.6
|
Slco1b2
|
solute carrier organic anion transporter family, member 1b2 |
| chr2_+_69219971 | 1.30 |
ENSMUST00000005364.5
ENSMUST00000112317.2 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr16_+_43235856 | 1.30 |
ENSMUST00000146708.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr15_-_81408261 | 1.30 |
ENSMUST00000057236.3
|
Dnajb7
|
DnaJ (Hsp40) homolog, subfamily B, member 7 |
| chr8_+_45627946 | 1.30 |
ENSMUST00000145458.1
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chrX_+_107149454 | 1.30 |
ENSMUST00000125676.1
ENSMUST00000180182.1 |
A630033H20Rik
|
RIKEN cDNA A630033H20 gene |
| chr5_+_107497762 | 1.28 |
ENSMUST00000152474.1
ENSMUST00000060553.7 |
A830010M20Rik
|
RIKEN cDNA A830010M20 gene |
| chr9_-_71896047 | 1.28 |
ENSMUST00000184448.1
|
Tcf12
|
transcription factor 12 |
| chr8_-_61902669 | 1.26 |
ENSMUST00000121785.1
ENSMUST00000034057.7 |
Palld
|
palladin, cytoskeletal associated protein |
| chr9_+_32116040 | 1.26 |
ENSMUST00000174641.1
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr5_-_51567717 | 1.26 |
ENSMUST00000127135.1
ENSMUST00000151104.1 |
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr9_+_54980880 | 1.25 |
ENSMUST00000093844.3
|
Chrna5
|
cholinergic receptor, nicotinic, alpha polypeptide 5 |
| chr17_+_79626669 | 1.24 |
ENSMUST00000086570.1
|
4921513D11Rik
|
RIKEN cDNA 4921513D11 gene |
| chr19_-_12765447 | 1.24 |
ENSMUST00000112933.1
|
Cntf
|
ciliary neurotrophic factor |
| chr10_+_87859062 | 1.23 |
ENSMUST00000095360.4
|
Igf1
|
insulin-like growth factor 1 |
| chr3_-_152193803 | 1.22 |
ENSMUST00000050073.6
|
Dnajb4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chrX_-_109013389 | 1.22 |
ENSMUST00000033597.8
|
Hmgn5
|
high-mobility group nucleosome binding domain 5 |
| chr4_+_48663502 | 1.21 |
ENSMUST00000030033.4
|
Murc
|
muscle-related coiled-coil protein |
| chr1_+_127729405 | 1.21 |
ENSMUST00000038006.6
|
Acmsd
|
amino carboxymuconate semialdehyde decarboxylase |
| chr16_-_23988852 | 1.21 |
ENSMUST00000023151.5
|
Bcl6
|
B cell leukemia/lymphoma 6 |
| chr9_-_13245741 | 1.21 |
ENSMUST00000110582.2
|
Jrkl
|
jerky homolog-like (mouse) |
| chr6_+_34709610 | 1.21 |
ENSMUST00000031775.6
|
Cald1
|
caldesmon 1 |
| chr2_-_7395968 | 1.21 |
ENSMUST00000002176.6
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr18_-_84086379 | 1.20 |
ENSMUST00000060303.8
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr10_+_26772477 | 1.20 |
ENSMUST00000039557.7
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr10_+_69925484 | 1.19 |
ENSMUST00000182692.1
ENSMUST00000092433.5 |
Ank3
|
ankyrin 3, epithelial |
| chr9_-_60838200 | 1.19 |
ENSMUST00000063858.7
|
Gm9869
|
predicted gene 9869 |
| chr6_-_99096196 | 1.19 |
ENSMUST00000175886.1
|
Foxp1
|
forkhead box P1 |
| chr8_+_85432686 | 1.19 |
ENSMUST00000180883.1
|
1700051O22Rik
|
RIKEN cDNA 1700051O22 Gene |
| chr11_+_23306910 | 1.18 |
ENSMUST00000137823.1
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr2_-_116067391 | 1.17 |
ENSMUST00000140185.1
|
2700033N17Rik
|
RIKEN cDNA 2700033N17 gene |
| chr19_+_55894508 | 1.17 |
ENSMUST00000142291.1
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 11.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.9 | 3.8 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.9 | 7.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.9 | 6.8 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.8 | 6.5 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.8 | 6.8 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.7 | 2.2 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.7 | 2.1 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) epicardium-derived cardiac vascular smooth muscle cell differentiation(GO:0060983) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 0.7 | 3.4 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.7 | 7.2 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.6 | 2.9 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.6 | 1.7 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.6 | 1.7 | GO:0002358 | B cell homeostatic proliferation(GO:0002358) |
| 0.6 | 7.7 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.5 | 3.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.5 | 3.3 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.5 | 2.7 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.5 | 2.1 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.5 | 1.6 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.5 | 1.6 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.5 | 3.1 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.5 | 2.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.5 | 2.6 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.5 | 8.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.5 | 3.5 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.5 | 1.5 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.5 | 4.5 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.5 | 1.5 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.5 | 1.9 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.5 | 1.4 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.5 | 1.9 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.5 | 6.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.5 | 1.8 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.5 | 0.9 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.5 | 1.8 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.4 | 3.5 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.4 | 3.0 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.4 | 2.6 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.4 | 2.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.4 | 2.0 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.4 | 1.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.4 | 2.0 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.4 | 1.6 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.4 | 2.7 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.4 | 1.2 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.4 | 1.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.4 | 1.5 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.4 | 3.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.4 | 3.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.4 | 1.5 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.4 | 2.3 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.4 | 0.4 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) |
| 0.4 | 1.1 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.4 | 1.5 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.4 | 1.1 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.4 | 2.6 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.4 | 4.0 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.3 | 2.4 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.3 | 1.7 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.3 | 1.0 | GO:1904835 | vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 0.3 | 2.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.3 | 1.3 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.3 | 1.6 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.3 | 1.6 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.3 | 0.6 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.3 | 0.9 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.3 | 1.9 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.3 | 0.9 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.3 | 0.9 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.3 | 3.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 1.2 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.3 | 0.9 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.3 | 1.7 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.3 | 1.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.3 | 0.3 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.3 | 1.4 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.3 | 1.1 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.3 | 1.6 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.3 | 3.2 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.3 | 1.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.3 | 0.3 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.3 | 1.0 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.3 | 0.8 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.3 | 1.8 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 1.0 | GO:0007494 | midgut development(GO:0007494) |
| 0.2 | 1.5 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 4.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.2 | 0.9 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.2 | 2.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 0.2 | GO:0046619 | optic placode formation(GO:0001743) optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.2 | 0.9 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.2 | 0.9 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.2 | 1.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 2.0 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.2 | 1.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 1.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 1.5 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 1.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 0.6 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.2 | 0.4 | GO:0060003 | copper ion export(GO:0060003) |
| 0.2 | 0.6 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.2 | 6.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 8.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 0.8 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.2 | 5.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 0.8 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 1.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.2 | 0.6 | GO:1904978 | regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 0.2 | 1.0 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.2 | 0.6 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.2 | 0.6 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.2 | 3.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.2 | 1.0 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.2 | 1.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 0.8 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) lung ciliated cell differentiation(GO:0061141) regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 0.4 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.2 | 2.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.2 | 5.4 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.2 | 0.6 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.2 | 0.8 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.2 | 0.8 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.2 | 0.6 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.2 | 0.8 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.2 | 0.4 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.2 | 2.6 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 | 0.6 | GO:0006930 | cap-dependent translational initiation(GO:0002191) substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.2 | 0.9 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.2 | 1.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.2 | 0.4 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.2 | 0.9 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.2 | 8.8 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 2.0 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.2 | 0.5 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.2 | 0.9 | GO:0072093 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.2 | 0.2 | GO:0035898 | parathyroid hormone secretion(GO:0035898) |
| 0.2 | 0.5 | GO:0090244 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) Wnt signaling pathway involved in somitogenesis(GO:0090244) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.2 | 0.2 | GO:0060982 | coronary artery morphogenesis(GO:0060982) |
| 0.2 | 0.7 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 1.9 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.2 | 1.7 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 | 0.9 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.2 | 1.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.2 | 1.0 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.2 | 0.5 | GO:0060618 | nipple development(GO:0060618) |
| 0.2 | 0.5 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 | 0.5 | GO:0072573 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.2 | 0.5 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.2 | 0.7 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 0.7 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.2 | 0.6 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.2 | 0.6 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.2 | 0.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.2 | 0.3 | GO:2000617 | positive regulation of histone H3-K14 acetylation(GO:0071442) positive regulation of histone H3-K9 acetylation(GO:2000617) |
| 0.2 | 0.8 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.2 | 0.8 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 0.8 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.2 | 2.9 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.2 | 0.6 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.2 | 1.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.2 | 0.9 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.2 | 0.5 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 1.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 0.5 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.2 | 2.1 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.1 | 0.9 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 1.0 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.9 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.4 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 2.0 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 1.6 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.1 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 0.1 | 1.1 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.1 | 0.3 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.1 | 0.8 | GO:0010255 | carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.1 | 0.6 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 0.6 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.7 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.4 | GO:2000229 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 0.1 | 0.5 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.1 | 0.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.9 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 3.5 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 0.5 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.8 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 1.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 1.6 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.9 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.1 | 0.9 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.4 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.2 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.1 | 0.7 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.7 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 2.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.2 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.4 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.1 | 3.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.5 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 0.8 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 1.2 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 | 0.3 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.1 | 1.0 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.3 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.6 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 1.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.9 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.6 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 | 0.4 | GO:1990401 | embryonic lung development(GO:1990401) |
| 0.1 | 0.4 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.1 | 0.4 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.1 | 1.1 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 2.5 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 0.4 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.1 | 0.3 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 0.3 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 | 0.4 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.1 | 1.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 0.1 | 0.5 | GO:0045356 | microglial cell activation involved in immune response(GO:0002282) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 1.6 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.1 | 0.8 | GO:0072176 | nephric duct development(GO:0072176) nephric duct morphogenesis(GO:0072178) |
| 0.1 | 0.7 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.1 | 1.3 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.2 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 | 2.9 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 0.6 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.4 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 1.7 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.4 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.2 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.1 | 0.3 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.1 | 0.3 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.1 | 0.3 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.1 | 1.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.4 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 0.8 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.7 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.3 | GO:0060353 | regulation of cell adhesion molecule production(GO:0060353) |
| 0.1 | 1.3 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 0.4 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.1 | 0.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.5 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 1.9 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.1 | 0.7 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.3 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) general adaptation syndrome(GO:0051866) |
| 0.1 | 0.9 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.1 | 0.8 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 1.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.5 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 1.7 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 1.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 0.5 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.4 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 0.8 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.6 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.5 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 1.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.5 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 0.5 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.6 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 1.4 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.7 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 1.8 | GO:0060044 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.1 | 1.7 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 0.5 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.2 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 0.8 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.1 | 0.8 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 | 1.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 3.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.7 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.7 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.2 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 0.1 | 5.2 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.4 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.1 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.1 | 4.9 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.5 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.1 | 0.6 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 0.4 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.2 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 0.3 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.1 | 3.3 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 1.8 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.4 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.5 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.1 | 0.6 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 1.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 2.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 2.0 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 1.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.2 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.4 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.1 | 3.6 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 0.2 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.1 | 0.1 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.1 | 0.6 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.5 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.1 | 1.2 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.1 | 0.1 | GO:0061144 | epithelial-mesenchymal cell signaling(GO:0060684) alveolar secondary septum development(GO:0061144) |
| 0.1 | 0.5 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 2.1 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.1 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.6 | GO:0006586 | tryptophan metabolic process(GO:0006568) indolalkylamine metabolic process(GO:0006586) |
| 0.1 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 1.2 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.4 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 0.2 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 0.3 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.2 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.6 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.1 | 0.3 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 1.2 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) response to glycoside(GO:1903416) |
| 0.1 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) positive regulation of neuromuscular junction development(GO:1904398) |
| 0.1 | 0.7 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.4 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 1.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.5 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.9 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.1 | 0.4 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.1 | 0.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 1.8 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.2 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 | 0.7 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.2 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.1 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.2 | GO:0014831 | gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.1 | 0.2 | GO:0043305 | negative regulation of mast cell activation involved in immune response(GO:0033007) negative regulation of mast cell degranulation(GO:0043305) |
| 0.1 | 5.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.5 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.1 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.1 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.5 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.1 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.3 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 0.6 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 0.1 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.0 | 1.0 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.6 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.4 | GO:1900364 | regulation of mRNA polyadenylation(GO:1900363) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 1.0 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.4 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.5 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.6 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.2 | GO:0045423 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of interleukin-8 biosynthetic process(GO:0045414) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 2.9 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 0.0 | 1.0 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.9 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.3 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.6 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.4 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.3 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.2 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 1.0 | GO:0002504 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.3 | GO:0046601 | positive regulation of centriole replication(GO:0046601) de novo centriole assembly(GO:0098535) |
| 0.0 | 0.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.6 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.9 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.3 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.2 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 3.6 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.4 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 0.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.0 | GO:0021767 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) |
| 0.0 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.6 | GO:0033866 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0021594 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.3 | GO:0007350 | blastoderm segmentation(GO:0007350) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.1 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.0 | 0.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.3 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.1 | GO:1902913 | positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.2 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.4 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.4 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.4 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.4 | GO:0034377 | plasma lipoprotein particle assembly(GO:0034377) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 1.2 | GO:0050879 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.0 | 0.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 1.2 | GO:0051058 | negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.4 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.7 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.2 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 1.4 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.8 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.2 | GO:0042997 | protein depalmitoylation(GO:0002084) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:1990927 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.2 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.5 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 1.1 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 1.3 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.3 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 3.0 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.8 | GO:0030858 | positive regulation of epithelial cell differentiation(GO:0030858) |
| 0.0 | 0.3 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.2 | GO:0051197 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 | 0.2 | GO:0097369 | sodium ion import(GO:0097369) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.3 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.8 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.8 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.8 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 1.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 2.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.2 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.5 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.1 | GO:0060767 | epithelial cell proliferation involved in prostate gland development(GO:0060767) regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) |
| 0.0 | 0.0 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.2 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.0 | 0.2 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 1.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 1.9 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.3 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0072498 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 2.0 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.0 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.0 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.0 | 0.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.4 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.5 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.4 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.0 | GO:0044857 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.0 | 0.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.0 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.0 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.3 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.3 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.2 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.3 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 1.2 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.5 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.1 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.2 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.6 | 11.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.5 | 4.7 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.4 | 4.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.4 | 1.3 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.4 | 1.6 | GO:0008623 | CHRAC(GO:0008623) |
| 0.4 | 2.0 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.4 | 4.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 1.4 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.3 | 4.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 1.3 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.3 | 1.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 0.9 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.3 | 2.6 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) insulin-like growth factor ternary complex(GO:0042567) |
| 0.3 | 0.5 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.3 | 7.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 7.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 0.2 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.2 | 4.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 0.8 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 1.0 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.2 | 2.7 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.2 | 4.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 1.1 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.2 | 0.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 4.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 0.9 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 1.0 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 0.8 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 1.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 3.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 1.4 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.4 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.9 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.7 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 6.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 6.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 8.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.8 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.3 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 1.8 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 2.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.4 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 1.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 6.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 3.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.8 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.1 | 1.7 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 1.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 2.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.9 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.7 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 0.7 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 1.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 1.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.6 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.7 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.5 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 1.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.3 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 9.5 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 1.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 2.3 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.4 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 0.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.4 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.4 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.7 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 0.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 0.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.7 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 1.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.6 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 0.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.7 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.8 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.1 | 4.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.2 | GO:0030934 | collagen type XII trimer(GO:0005595) anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 2.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 2.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.4 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 3.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 3.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.8 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 7.6 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 1.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 6.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.8 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 5.2 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 4.4 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 8.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 8.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 4.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 2.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 6.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.3 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 1.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 1.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 23.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.1 | GO:1990131 | Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.6 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 5.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 1.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.4 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.9 | 3.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.8 | 0.8 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.8 | 3.0 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.7 | 2.2 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.6 | 1.8 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.6 | 2.4 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.6 | 1.7 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.5 | 5.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.5 | 2.1 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.5 | 8.0 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.5 | 2.3 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.5 | 2.8 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.4 | 1.8 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.4 | 1.7 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.4 | 0.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.4 | 2.0 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.4 | 1.6 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.4 | 1.2 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.4 | 1.9 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.4 | 1.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.4 | 1.1 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.4 | 1.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.3 | 6.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.3 | 3.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 1.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.3 | 6.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.3 | 2.0 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.3 | 10.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.3 | 1.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.3 | 1.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 0.8 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.3 | 7.8 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 1.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 2.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 0.7 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.2 | 1.5 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 0.7 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.2 | 1.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 2.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 0.2 | GO:0071820 | N-box binding(GO:0071820) |
| 0.2 | 0.7 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 1.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 1.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.9 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 1.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 1.4 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 0.6 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.2 | 1.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 1.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 2.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 2.8 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 2.8 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 1.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.2 | 0.2 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.2 | 2.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 1.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 1.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 0.6 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.2 | 3.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 0.6 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.2 | 2.7 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.2 | 1.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 0.4 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.2 | 3.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 0.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 4.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.2 | 0.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 6.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 2.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 0.5 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.2 | 1.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 0.3 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.2 | 1.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.2 | 3.5 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 0.7 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 1.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 1.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 0.5 | GO:0016520 | gonadotropin-releasing hormone receptor activity(GO:0004968) growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.2 | 0.5 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.2 | 0.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.7 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 3.7 | GO:0005402 | cation:sugar symporter activity(GO:0005402) |
| 0.1 | 0.4 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 1.6 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 9.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 9.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.7 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 7.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 1.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.0 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 3.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 8.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.7 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.1 | 2.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.4 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 1.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.4 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.1 | 0.9 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 2.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.6 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 3.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.4 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 1.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.5 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.1 | 0.9 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.4 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 2.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.5 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.6 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.6 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.6 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 0.5 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 0.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 1.9 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.3 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.1 | 1.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.5 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 1.6 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 1.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.4 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 1.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 3.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 3.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 2.3 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 0.7 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 3.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.0 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.6 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.3 | GO:0038100 | nodal binding(GO:0038100) |
| 0.1 | 0.6 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.4 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 1.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.3 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.6 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 0.9 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.2 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 0.6 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 1.2 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.2 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.1 | 0.8 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 0.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 1.6 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 1.0 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 1.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.4 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 1.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 1.2 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 0.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 2.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.5 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 1.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 1.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.2 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 0.2 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 1.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.6 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.2 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.1 | 1.1 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 1.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 6.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 1.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 1.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 2.1 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 1.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 3.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 2.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 5.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.4 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.6 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 1.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 1.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.4 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 1.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 1.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0099589 | serotonin receptor activity(GO:0099589) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.4 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 1.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 3.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.2 | GO:0004321 | acyl-CoA ligase activity(GO:0003996) fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.3 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.3 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 8.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 5.8 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.6 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.1 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 3.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.5 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 5.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.0 | 1.0 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 5.3 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.0 | 0.0 | GO:0030977 | taurine binding(GO:0030977) |
| 0.0 | 0.1 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.1 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 16.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 12.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 13.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 15.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.2 | 6.9 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 8.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 5.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 4.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 4.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 0.5 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 0.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 3.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 1.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 0.9 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 3.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 2.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 3.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 5.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 6.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 5.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 3.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 0.7 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 1.3 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 2.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 2.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 4.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 2.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 1.5 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 1.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 1.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 2.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 2.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.5 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 7.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 3.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 4.0 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.3 | 8.0 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.3 | 4.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.3 | 2.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 3.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 9.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 2.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 4.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 6.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 2.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 2.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 2.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 0.6 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.1 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 12.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 2.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 9.9 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 0.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 3.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 4.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 2.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 5.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 3.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 3.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 15.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 3.9 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.1 | 1.4 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 1.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 0.4 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 2.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.5 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 0.9 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 0.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.2 | REACTOME SIGNALING BY FGFR IN DISEASE | Genes involved in Signaling by FGFR in disease |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 1.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 2.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 2.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 2.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 2.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 2.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATION OF FGFR SIGNALING | Genes involved in Negative regulation of FGFR signaling |
| 0.0 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 3.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 1.1 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.1 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.0 | 0.6 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.7 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |