Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
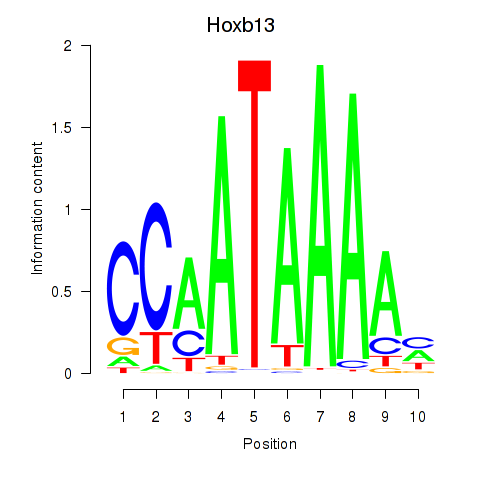
Results for Hoxb13
Z-value: 1.57
Transcription factors associated with Hoxb13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb13
|
ENSMUSG00000049604.3 | homeobox B13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb13 | mm10_v2_chr11_+_96194299_96194316 | -0.36 | 3.0e-02 | Click! |
Activity profile of Hoxb13 motif
Sorted Z-values of Hoxb13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_24576297 | 21.60 |
ENSMUST00000033953.7
ENSMUST00000121992.1 |
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr5_-_146009598 | 20.82 |
ENSMUST00000138870.1
ENSMUST00000068317.6 |
Cyp3a25
|
cytochrome P450, family 3, subfamily a, polypeptide 25 |
| chr5_+_146079254 | 20.34 |
ENSMUST00000035571.8
|
Cyp3a59
|
cytochrome P450, family 3, subfamily a, polypeptide 59 |
| chr4_-_61674094 | 12.84 |
ENSMUST00000098040.3
|
Mup18
|
major urinary protein 18 |
| chr19_-_8405060 | 11.42 |
ENSMUST00000064507.5
ENSMUST00000120540.1 ENSMUST00000096269.4 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr4_-_61303802 | 11.36 |
ENSMUST00000125461.1
|
Mup14
|
major urinary protein 14 |
| chr19_+_40089688 | 11.36 |
ENSMUST00000068094.6
ENSMUST00000080171.2 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr4_-_61519467 | 10.97 |
ENSMUST00000095051.5
ENSMUST00000107483.1 |
Mup16
|
major urinary protein 16 |
| chr4_-_61303998 | 10.69 |
ENSMUST00000071005.8
ENSMUST00000075206.5 |
Mup14
|
major urinary protein 14 |
| chr4_-_61439743 | 10.46 |
ENSMUST00000095049.4
|
Mup15
|
major urinary protein 15 |
| chr19_+_39287074 | 10.37 |
ENSMUST00000003137.8
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr4_-_61228271 | 10.35 |
ENSMUST00000072678.5
ENSMUST00000098042.3 |
Mup13
|
major urinary protein 13 |
| chr4_-_60070411 | 10.14 |
ENSMUST00000079697.3
ENSMUST00000125282.1 ENSMUST00000166098.1 |
Mup7
|
major urinary protein 7 |
| chr4_-_60662358 | 9.68 |
ENSMUST00000084544.4
ENSMUST00000098046.3 |
Mup11
|
major urinary protein 11 |
| chr19_+_39992424 | 9.53 |
ENSMUST00000049178.2
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr4_-_61595871 | 9.31 |
ENSMUST00000107484.1
|
Mup17
|
major urinary protein 17 |
| chr4_-_60501903 | 9.17 |
ENSMUST00000084548.4
ENSMUST00000103012.3 ENSMUST00000107499.3 |
Mup1
|
major urinary protein 1 |
| chr6_+_41302265 | 9.05 |
ENSMUST00000031913.4
|
Try4
|
trypsin 4 |
| chr4_-_60139857 | 8.99 |
ENSMUST00000107490.4
ENSMUST00000074700.2 |
Mup2
|
major urinary protein 2 |
| chr1_-_121332545 | 8.88 |
ENSMUST00000161068.1
|
Insig2
|
insulin induced gene 2 |
| chr12_-_104153846 | 8.86 |
ENSMUST00000085050.3
|
Serpina3c
|
serine (or cysteine) peptidase inhibitor, clade A, member 3C |
| chr13_-_23914998 | 8.61 |
ENSMUST00000021769.8
ENSMUST00000110407.2 |
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr4_-_60741275 | 8.37 |
ENSMUST00000117932.1
|
Mup12
|
major urinary protein 12 |
| chr4_-_60222580 | 8.31 |
ENSMUST00000095058.4
ENSMUST00000163931.1 |
Mup8
|
major urinary protein 8 |
| chr4_-_60421933 | 8.29 |
ENSMUST00000107506.2
ENSMUST00000122381.1 ENSMUST00000118759.1 ENSMUST00000122177.1 |
Mup9
|
major urinary protein 9 |
| chr4_-_60582152 | 8.07 |
ENSMUST00000098047.2
|
Mup10
|
major urinary protein 10 |
| chr15_-_75431745 | 8.05 |
ENSMUST00000096397.1
|
9030619P08Rik
|
RIKEN cDNA 9030619P08 gene |
| chr8_-_93279717 | 7.92 |
ENSMUST00000034178.8
|
Ces1f
|
carboxylesterase 1F |
| chr7_+_26808880 | 7.28 |
ENSMUST00000040944.7
|
Cyp2g1
|
cytochrome P450, family 2, subfamily g, polypeptide 1 |
| chr6_-_41314700 | 7.12 |
ENSMUST00000064324.5
|
Try5
|
trypsin 5 |
| chr5_-_87337165 | 7.03 |
ENSMUST00000031195.2
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr1_-_121332571 | 6.96 |
ENSMUST00000071064.6
|
Insig2
|
insulin induced gene 2 |
| chr13_+_4434306 | 6.84 |
ENSMUST00000021630.8
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr1_-_150465563 | 6.81 |
ENSMUST00000164600.1
ENSMUST00000111902.2 ENSMUST00000111901.2 ENSMUST00000006171.9 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr6_-_85869128 | 6.76 |
ENSMUST00000045008.7
|
Cml2
|
camello-like 2 |
| chr19_+_12633303 | 6.75 |
ENSMUST00000044976.5
|
Glyat
|
glycine-N-acyltransferase |
| chr19_-_7802578 | 6.72 |
ENSMUST00000120522.1
ENSMUST00000065634.7 |
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr19_+_12633507 | 6.45 |
ENSMUST00000119960.1
|
Glyat
|
glycine-N-acyltransferase |
| chr17_-_31144271 | 6.36 |
ENSMUST00000024826.7
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr15_-_77411034 | 6.20 |
ENSMUST00000089452.5
ENSMUST00000081776.3 |
Apol9a
|
apolipoprotein L 9a |
| chr16_+_91269759 | 6.00 |
ENSMUST00000056882.5
|
Olig1
|
oligodendrocyte transcription factor 1 |
| chr7_+_44207307 | 5.74 |
ENSMUST00000077354.4
|
Klk1b4
|
kallikrein 1-related pepidase b4 |
| chr1_-_193264006 | 5.71 |
ENSMUST00000161737.1
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr7_+_132610620 | 5.69 |
ENSMUST00000033241.5
|
Lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr8_+_45069137 | 5.69 |
ENSMUST00000067984.7
|
Mtnr1a
|
melatonin receptor 1A |
| chr4_-_62054112 | 5.68 |
ENSMUST00000074018.3
|
Mup20
|
major urinary protein 20 |
| chr4_-_115496129 | 5.56 |
ENSMUST00000030487.2
|
Cyp4a14
|
cytochrome P450, family 4, subfamily a, polypeptide 14 |
| chr15_+_77729091 | 5.54 |
ENSMUST00000109775.2
|
Apol9b
|
apolipoprotein L 9b |
| chr15_+_6445320 | 5.36 |
ENSMUST00000022749.9
|
C9
|
complement component 9 |
| chr13_-_41847482 | 5.02 |
ENSMUST00000072012.3
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr7_+_44198191 | 4.96 |
ENSMUST00000085450.2
|
Klk1b3
|
kallikrein 1-related peptidase b3 |
| chr4_+_115518264 | 4.88 |
ENSMUST00000058785.3
ENSMUST00000094886.3 |
Cyp4a10
|
cytochrome P450, family 4, subfamily a, polypeptide 10 |
| chr13_-_41847599 | 4.88 |
ENSMUST00000179758.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr5_-_87091150 | 4.86 |
ENSMUST00000154455.1
|
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr4_-_49383576 | 4.75 |
ENSMUST00000107698.1
|
Acnat2
|
acyl-coenzyme A amino acid N-acyltransferase 2 |
| chr3_+_129836729 | 4.67 |
ENSMUST00000077918.5
|
Cfi
|
complement component factor i |
| chr8_+_70083509 | 4.66 |
ENSMUST00000007738.9
|
Hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr1_+_88095054 | 4.59 |
ENSMUST00000150634.1
ENSMUST00000058237.7 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr17_+_31433054 | 4.49 |
ENSMUST00000136384.1
|
Pde9a
|
phosphodiesterase 9A |
| chr4_-_108118504 | 4.46 |
ENSMUST00000149106.1
|
Scp2
|
sterol carrier protein 2, liver |
| chr19_-_8218832 | 4.41 |
ENSMUST00000113298.2
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr9_-_51328898 | 4.41 |
ENSMUST00000039959.4
|
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr4_+_20007938 | 4.35 |
ENSMUST00000125799.1
ENSMUST00000121491.1 |
Ttpa
|
tocopherol (alpha) transfer protein |
| chr4_+_141368116 | 4.28 |
ENSMUST00000006380.4
|
Fam131c
|
family with sequence similarity 131, member C |
| chrM_+_3906 | 4.21 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr11_-_113710017 | 4.19 |
ENSMUST00000018871.1
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr10_+_84756055 | 4.18 |
ENSMUST00000060397.6
|
Rfx4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr1_+_13668739 | 4.16 |
ENSMUST00000088542.3
|
Xkr9
|
X Kell blood group precursor related family member 9 homolog |
| chr1_-_130661584 | 4.14 |
ENSMUST00000137276.2
|
C4bp
|
complement component 4 binding protein |
| chr7_-_68275098 | 4.12 |
ENSMUST00000135564.1
|
Gm16157
|
predicted gene 16157 |
| chr7_-_12998172 | 4.01 |
ENSMUST00000120903.1
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr1_-_130661613 | 4.01 |
ENSMUST00000027657.7
|
C4bp
|
complement component 4 binding protein |
| chr5_-_145879857 | 4.00 |
ENSMUST00000035918.7
|
Cyp3a11
|
cytochrome P450, family 3, subfamily a, polypeptide 11 |
| chr7_-_139582790 | 3.96 |
ENSMUST00000106095.2
|
Nkx6-2
|
NK6 homeobox 2 |
| chr11_-_11898092 | 3.94 |
ENSMUST00000178704.1
|
Ddc
|
dopa decarboxylase |
| chr14_-_51913393 | 3.92 |
ENSMUST00000004673.7
ENSMUST00000111632.3 |
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr14_+_65969714 | 3.89 |
ENSMUST00000153460.1
|
Clu
|
clusterin |
| chr16_+_22951072 | 3.87 |
ENSMUST00000023590.8
|
Hrg
|
histidine-rich glycoprotein |
| chr6_+_42245907 | 3.85 |
ENSMUST00000031897.5
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr8_-_109579056 | 3.84 |
ENSMUST00000074898.6
|
Hp
|
haptoglobin |
| chr7_+_119607014 | 3.83 |
ENSMUST00000126367.1
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr3_+_19985612 | 3.82 |
ENSMUST00000172860.1
|
Cp
|
ceruloplasmin |
| chr5_+_114175889 | 3.71 |
ENSMUST00000146841.1
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr5_+_137981512 | 3.65 |
ENSMUST00000035390.5
|
Azgp1
|
alpha-2-glycoprotein 1, zinc |
| chrX_+_59999436 | 3.55 |
ENSMUST00000033477.4
|
F9
|
coagulation factor IX |
| chr8_+_95352258 | 3.46 |
ENSMUST00000034243.5
|
Mmp15
|
matrix metallopeptidase 15 |
| chr6_+_41354105 | 3.45 |
ENSMUST00000072103.5
|
Try10
|
trypsin 10 |
| chr19_-_11081088 | 3.42 |
ENSMUST00000025636.6
|
Ms4a8a
|
membrane-spanning 4-domains, subfamily A, member 8A |
| chr14_+_65968483 | 3.41 |
ENSMUST00000022616.6
|
Clu
|
clusterin |
| chr8_-_117671526 | 3.35 |
ENSMUST00000037955.7
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr8_-_105933832 | 3.34 |
ENSMUST00000034368.6
|
Ctrl
|
chymotrypsin-like |
| chr4_-_108217897 | 3.33 |
ENSMUST00000106690.1
ENSMUST00000043793.6 |
Zyg11a
|
zyg-11 family member A, cell cycle regulator |
| chr14_-_47189406 | 3.30 |
ENSMUST00000089959.6
|
Gch1
|
GTP cyclohydrolase 1 |
| chr13_-_41847626 | 3.29 |
ENSMUST00000121404.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr16_+_56204313 | 3.22 |
ENSMUST00000160116.1
ENSMUST00000069936.7 |
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr6_-_85933379 | 3.21 |
ENSMUST00000162660.1
|
Nat8b
|
N-acetyltransferase 8B |
| chr5_+_114923234 | 3.21 |
ENSMUST00000031540.4
ENSMUST00000112143.3 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chrX_-_100594860 | 3.19 |
ENSMUST00000053373.1
|
P2ry4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr4_+_144893127 | 3.15 |
ENSMUST00000142808.1
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr5_-_113081579 | 3.12 |
ENSMUST00000131708.1
ENSMUST00000117143.1 ENSMUST00000119627.1 |
Crybb3
|
crystallin, beta B3 |
| chr16_-_23988852 | 3.08 |
ENSMUST00000023151.5
|
Bcl6
|
B cell leukemia/lymphoma 6 |
| chr4_+_99030946 | 3.07 |
ENSMUST00000030280.6
|
Angptl3
|
angiopoietin-like 3 |
| chr5_-_87424201 | 3.06 |
ENSMUST00000072818.5
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr12_-_98577940 | 3.03 |
ENSMUST00000110113.1
|
Kcnk10
|
potassium channel, subfamily K, member 10 |
| chr12_+_108334341 | 3.03 |
ENSMUST00000021684.4
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr10_-_107123585 | 3.02 |
ENSMUST00000165067.1
ENSMUST00000044668.4 |
Acss3
|
acyl-CoA synthetase short-chain family member 3 |
| chr1_+_160978576 | 3.01 |
ENSMUST00000064725.5
|
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr8_-_25038875 | 2.98 |
ENSMUST00000084031.4
|
Htra4
|
HtrA serine peptidase 4 |
| chr19_+_30232921 | 2.97 |
ENSMUST00000025797.5
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chr2_-_32451396 | 2.95 |
ENSMUST00000028160.8
ENSMUST00000113310.2 |
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr19_-_39649046 | 2.93 |
ENSMUST00000067328.6
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr11_+_94211431 | 2.91 |
ENSMUST00000041589.5
|
Tob1
|
transducer of ErbB-2.1 |
| chr8_-_5105232 | 2.90 |
ENSMUST00000023835.1
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr5_-_45450221 | 2.90 |
ENSMUST00000015950.5
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr9_+_107340593 | 2.89 |
ENSMUST00000042581.2
|
6430571L13Rik
|
RIKEN cDNA 6430571L13 gene |
| chr3_+_62338344 | 2.88 |
ENSMUST00000079300.6
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr8_-_41215146 | 2.85 |
ENSMUST00000034003.4
|
Fgl1
|
fibrinogen-like protein 1 |
| chr6_+_124512615 | 2.84 |
ENSMUST00000068593.7
|
C1ra
|
complement component 1, r subcomponent A |
| chr2_+_25054396 | 2.82 |
ENSMUST00000102931.4
ENSMUST00000074422.7 ENSMUST00000132172.1 ENSMUST00000114388.1 ENSMUST00000114386.1 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr7_+_127800604 | 2.82 |
ENSMUST00000046863.5
ENSMUST00000106272.1 ENSMUST00000139068.1 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr11_-_73324616 | 2.81 |
ENSMUST00000021119.2
|
Aspa
|
aspartoacylase |
| chr5_-_77095225 | 2.80 |
ENSMUST00000120827.2
ENSMUST00000113453.2 |
Hopx
|
HOP homeobox |
| chr5_-_87538188 | 2.80 |
ENSMUST00000031199.4
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr4_+_102570065 | 2.77 |
ENSMUST00000097950.2
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr11_+_97029925 | 2.73 |
ENSMUST00000021249.4
|
Scrn2
|
secernin 2 |
| chr3_+_19957088 | 2.73 |
ENSMUST00000108328.1
|
Cp
|
ceruloplasmin |
| chr14_-_68533689 | 2.72 |
ENSMUST00000022640.7
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr9_-_50555170 | 2.69 |
ENSMUST00000119103.1
|
Bco2
|
beta-carotene oxygenase 2 |
| chr10_-_115362191 | 2.67 |
ENSMUST00000092170.5
|
Tmem19
|
transmembrane protein 19 |
| chr18_-_38209762 | 2.67 |
ENSMUST00000057185.6
|
Pcdh1
|
protocadherin 1 |
| chr5_-_87569023 | 2.65 |
ENSMUST00000113314.2
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr6_+_124304646 | 2.65 |
ENSMUST00000112541.2
ENSMUST00000032234.2 |
Cd163
|
CD163 antigen |
| chr9_-_44799179 | 2.64 |
ENSMUST00000114705.1
ENSMUST00000002100.7 |
Tmem25
|
transmembrane protein 25 |
| chr13_+_34734837 | 2.64 |
ENSMUST00000039605.6
|
Fam50b
|
family with sequence similarity 50, member B |
| chr15_+_31224371 | 2.63 |
ENSMUST00000044524.9
|
Dap
|
death-associated protein |
| chr16_-_31314804 | 2.60 |
ENSMUST00000115230.1
ENSMUST00000130560.1 |
Apod
|
apolipoprotein D |
| chr18_-_3281752 | 2.59 |
ENSMUST00000140332.1
ENSMUST00000147138.1 |
Crem
|
cAMP responsive element modulator |
| chr5_-_87591582 | 2.59 |
ENSMUST00000031201.7
|
Sult1e1
|
sulfotransferase family 1E, member 1 |
| chr13_+_24845122 | 2.58 |
ENSMUST00000006893.8
|
D130043K22Rik
|
RIKEN cDNA D130043K22 gene |
| chr1_-_162866502 | 2.57 |
ENSMUST00000046049.7
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr15_-_39857459 | 2.54 |
ENSMUST00000022915.3
ENSMUST00000110306.1 |
Dpys
|
dihydropyrimidinase |
| chr3_+_19957037 | 2.53 |
ENSMUST00000091309.5
ENSMUST00000108329.1 ENSMUST00000003714.6 |
Cp
|
ceruloplasmin |
| chr11_-_116189542 | 2.51 |
ENSMUST00000148601.1
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr2_-_18048784 | 2.51 |
ENSMUST00000142856.1
|
Skida1
|
SKI/DACH domain containing 1 |
| chr11_-_4118778 | 2.50 |
ENSMUST00000003681.7
|
Sec14l2
|
SEC14-like 2 (S. cerevisiae) |
| chr16_+_13940630 | 2.49 |
ENSMUST00000141971.1
ENSMUST00000124947.1 ENSMUST00000023360.7 ENSMUST00000143697.1 |
Mpv17l
|
Mpv17 transgene, kidney disease mutant-like |
| chr3_+_19957240 | 2.49 |
ENSMUST00000108325.2
|
Cp
|
ceruloplasmin |
| chr3_+_142701067 | 2.48 |
ENSMUST00000044392.4
|
Ccbl2
|
cysteine conjugate-beta lyase 2 |
| chr15_-_31453564 | 2.47 |
ENSMUST00000110408.1
|
Ropn1l
|
ropporin 1-like |
| chr7_-_12998140 | 2.47 |
ENSMUST00000032539.7
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr11_+_101665541 | 2.47 |
ENSMUST00000039388.2
|
Arl4d
|
ADP-ribosylation factor-like 4D |
| chr17_-_91092715 | 2.46 |
ENSMUST00000160800.2
ENSMUST00000159778.1 ENSMUST00000160844.3 |
Nrxn1
|
neurexin I |
| chr3_+_138313279 | 2.43 |
ENSMUST00000013455.6
ENSMUST00000106247.1 |
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr2_-_91195035 | 2.42 |
ENSMUST00000111356.1
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr15_-_77399086 | 2.41 |
ENSMUST00000175919.1
ENSMUST00000176074.1 |
Apol7a
|
apolipoprotein L 7a |
| chr9_-_106247730 | 2.41 |
ENSMUST00000112524.2
ENSMUST00000074082.6 |
Alas1
|
aminolevulinic acid synthase 1 |
| chr14_-_37048957 | 2.40 |
ENSMUST00000022338.5
|
Rgr
|
retinal G protein coupled receptor |
| chr5_-_45450143 | 2.40 |
ENSMUST00000154962.1
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr6_+_71199827 | 2.39 |
ENSMUST00000067492.7
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr16_-_46155077 | 2.39 |
ENSMUST00000059524.5
|
Gm4737
|
predicted gene 4737 |
| chr10_+_41490436 | 2.38 |
ENSMUST00000105507.3
|
Ppil6
|
peptidylprolyl isomerase (cyclophilin)-like 6 |
| chr1_-_97128249 | 2.37 |
ENSMUST00000027569.7
|
Slco6c1
|
solute carrier organic anion transporter family, member 6c1 |
| chr2_-_91195097 | 2.36 |
ENSMUST00000002177.2
ENSMUST00000111354.1 |
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr5_-_88526496 | 2.35 |
ENSMUST00000164073.1
|
Igj
|
immunoglobulin joining chain |
| chr8_-_105943382 | 2.34 |
ENSMUST00000038896.7
|
Lcat
|
lecithin cholesterol acyltransferase |
| chr1_-_69108039 | 2.34 |
ENSMUST00000121473.1
|
Erbb4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) |
| chr2_-_91236877 | 2.33 |
ENSMUST00000111352.1
|
Ddb2
|
damage specific DNA binding protein 2 |
| chr10_+_23851454 | 2.33 |
ENSMUST00000020190.7
|
Vnn3
|
vanin 3 |
| chr2_-_86347764 | 2.33 |
ENSMUST00000099894.2
|
Olfr1055
|
olfactory receptor 1055 |
| chr11_-_110095937 | 2.33 |
ENSMUST00000106664.3
ENSMUST00000046223.7 ENSMUST00000106662.1 |
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr17_+_34197715 | 2.33 |
ENSMUST00000173441.1
ENSMUST00000025196.8 |
Psmb8
|
proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) |
| chr2_+_177508570 | 2.30 |
ENSMUST00000108940.2
|
Gm14403
|
predicted gene 14403 |
| chr2_-_144550777 | 2.30 |
ENSMUST00000028915.5
|
Rbbp9
|
retinoblastoma binding protein 9 |
| chr15_-_34495180 | 2.29 |
ENSMUST00000022946.5
|
Hrsp12
|
heat-responsive protein 12 |
| chr4_+_20008357 | 2.29 |
ENSMUST00000117632.1
ENSMUST00000098244.1 |
Ttpa
|
tocopherol (alpha) transfer protein |
| chr18_-_31820413 | 2.28 |
ENSMUST00000165131.2
|
Gm6665
|
predicted gene 6665 |
| chr1_+_185332143 | 2.26 |
ENSMUST00000027916.6
ENSMUST00000151769.1 ENSMUST00000110965.1 |
Bpnt1
|
bisphosphate 3'-nucleotidase 1 |
| chrX_-_75380041 | 2.26 |
ENSMUST00000114085.2
|
F8
|
coagulation factor VIII |
| chr8_-_94696223 | 2.24 |
ENSMUST00000034227.4
|
Pllp
|
plasma membrane proteolipid |
| chr9_+_7692086 | 2.20 |
ENSMUST00000018767.7
|
Mmp7
|
matrix metallopeptidase 7 |
| chr8_+_45507768 | 2.17 |
ENSMUST00000067065.7
ENSMUST00000098788.3 ENSMUST00000067107.7 ENSMUST00000171337.2 ENSMUST00000138049.1 ENSMUST00000141039.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr1_-_36244245 | 2.16 |
ENSMUST00000046875.7
|
Uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr2_-_69342600 | 2.16 |
ENSMUST00000102709.1
ENSMUST00000102710.3 ENSMUST00000180142.1 |
Abcb11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr8_+_45069374 | 2.15 |
ENSMUST00000130141.1
|
Mtnr1a
|
melatonin receptor 1A |
| chr12_-_103657095 | 2.15 |
ENSMUST00000152517.1
|
Serpina6
|
serine (or cysteine) peptidase inhibitor, clade A, member 6 |
| chr15_+_32920723 | 2.14 |
ENSMUST00000022871.5
|
Sdc2
|
syndecan 2 |
| chr11_-_21371143 | 2.13 |
ENSMUST00000060895.5
|
Ugp2
|
UDP-glucose pyrophosphorylase 2 |
| chr6_+_40628824 | 2.13 |
ENSMUST00000071535.6
|
Mgam
|
maltase-glucoamylase |
| chrX_+_139563316 | 2.12 |
ENSMUST00000113027.1
|
Rnf128
|
ring finger protein 128 |
| chr7_-_80403315 | 2.12 |
ENSMUST00000147150.1
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr12_+_85288591 | 2.11 |
ENSMUST00000059341.4
|
Zc2hc1c
|
zinc finger, C2HC-type containing 1C |
| chr10_-_81427114 | 2.11 |
ENSMUST00000078185.7
ENSMUST00000020461.8 ENSMUST00000105321.3 |
Nfic
|
nuclear factor I/C |
| chr13_+_93674403 | 2.10 |
ENSMUST00000048001.6
|
Dmgdh
|
dimethylglycine dehydrogenase precursor |
| chr9_+_43829963 | 2.08 |
ENSMUST00000180221.1
|
Gm3898
|
predicted gene 3898 |
| chr8_-_121944886 | 2.07 |
ENSMUST00000057653.7
|
Car5a
|
carbonic anhydrase 5a, mitochondrial |
| chr4_+_148140699 | 2.07 |
ENSMUST00000140049.1
ENSMUST00000105707.1 |
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr11_-_61378052 | 2.06 |
ENSMUST00000010267.3
|
Slc47a1
|
solute carrier family 47, member 1 |
| chr19_-_34879452 | 2.06 |
ENSMUST00000036584.5
|
Pank1
|
pantothenate kinase 1 |
| chr1_+_88200601 | 2.06 |
ENSMUST00000049289.8
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chrX_+_142226765 | 2.04 |
ENSMUST00000112916.2
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.9 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 2.2 | 24.1 | GO:0006569 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 2.1 | 6.3 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 1.8 | 5.5 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 1.7 | 18.8 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 1.7 | 6.6 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 1.6 | 4.9 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 1.6 | 6.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 1.6 | 22.5 | GO:0015747 | urate transport(GO:0015747) |
| 1.6 | 4.8 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 1.5 | 4.6 | GO:0046271 | coumarin metabolic process(GO:0009804) phenylpropanoid catabolic process(GO:0046271) |
| 1.3 | 44.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.3 | 3.9 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 1.3 | 3.8 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 1.3 | 5.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 1.2 | 3.7 | GO:2001293 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 1.2 | 7.3 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.1 | 4.5 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 1.1 | 3.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 1.0 | 17.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 1.0 | 3.9 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.0 | 8.6 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.9 | 5.7 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.9 | 6.6 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.9 | 5.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.9 | 0.9 | GO:0002465 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) |
| 0.9 | 3.5 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.9 | 2.6 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.9 | 6.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.8 | 2.5 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.8 | 1.7 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.8 | 4.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.8 | 2.3 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.8 | 2.3 | GO:0019265 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-alanine catabolic process(GO:0042853) |
| 0.7 | 4.4 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.7 | 2.8 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.7 | 2.0 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.7 | 2.0 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.7 | 5.3 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.7 | 2.0 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.6 | 1.9 | GO:0010986 | GPI anchor release(GO:0006507) response to iron(II) ion(GO:0010040) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.6 | 1.2 | GO:0003130 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.6 | 2.5 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.6 | 3.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.6 | 1.8 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.6 | 2.5 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.6 | 3.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.6 | 2.4 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.6 | 9.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.6 | 1.7 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.6 | 9.1 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.6 | 4.0 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.6 | 1.7 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.6 | 3.9 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.6 | 1.7 | GO:0001193 | maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.6 | 2.2 | GO:0002780 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 0.5 | 2.7 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.5 | 1.6 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.5 | 1.6 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.5 | 5.8 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.5 | 2.7 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.5 | 2.1 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.5 | 4.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.5 | 2.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.5 | 2.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.5 | 1.5 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.5 | 1.5 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.5 | 1.5 | GO:0072343 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 0.5 | 1.5 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.5 | 1.4 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.5 | 1.9 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.5 | 8.9 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.5 | 2.3 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.5 | 2.8 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.5 | 1.4 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.5 | 1.4 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.5 | 7.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 1.3 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.4 | 11.9 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.4 | 3.5 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.4 | 1.3 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.4 | 1.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.4 | 2.5 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.4 | 2.5 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.4 | 1.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.4 | 0.8 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) |
| 0.4 | 1.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.4 | 3.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.4 | 1.6 | GO:0034759 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) |
| 0.4 | 1.1 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.4 | 1.1 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.4 | 1.5 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.4 | 15.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.4 | 1.4 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.4 | 0.4 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.4 | 2.8 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.3 | 1.4 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.3 | 4.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 5.0 | GO:0033866 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.3 | 0.7 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 4.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 1.3 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.3 | 1.2 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.3 | 1.5 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.3 | 2.7 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.3 | 2.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.3 | 1.5 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.3 | 1.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.3 | 1.7 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.3 | 2.2 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.3 | 1.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.3 | 0.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.3 | 0.8 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.3 | 3.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.3 | 3.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.3 | 2.9 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.3 | 2.9 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.3 | 2.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.3 | 1.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 4.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 2.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.2 | 2.9 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 1.5 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) ferrous iron transmembrane transport(GO:1903874) |
| 0.2 | 1.7 | GO:0042559 | folic acid-containing compound biosynthetic process(GO:0009396) pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.2 | 1.9 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.2 | 0.7 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 | 1.0 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.2 | 0.7 | GO:0031038 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.2 | 0.7 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.2 | 0.9 | GO:1902045 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.2 | 0.5 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.2 | 4.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 3.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 1.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 4.6 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.2 | 1.1 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.2 | 0.7 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.2 | 0.7 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.2 | 0.7 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 0.7 | GO:0002436 | immune complex clearance by monocytes and macrophages(GO:0002436) monocyte homeostasis(GO:0035702) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.2 | 3.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 0.9 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.2 | 27.0 | GO:0007586 | digestion(GO:0007586) |
| 0.2 | 1.5 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 | 2.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.2 | 2.0 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.2 | 0.6 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.2 | 3.8 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.2 | 0.6 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) |
| 0.2 | 0.8 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 | 1.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.2 | 2.5 | GO:0090205 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.2 | 1.2 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.2 | 0.6 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.2 | 8.9 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.2 | 3.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 0.8 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.2 | 0.6 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.2 | 1.0 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.2 | 1.5 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.2 | 1.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 4.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.2 | 1.7 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 0.6 | GO:0060618 | nipple development(GO:0060618) |
| 0.2 | 1.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 1.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.2 | 2.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.2 | 0.5 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.2 | 1.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 3.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 0.6 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.2 | 0.6 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.2 | 0.6 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 | 1.5 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.2 | 2.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.4 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 1.6 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.7 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 3.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 4.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 0.8 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 1.5 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 0.7 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.4 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 0.8 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 | 0.7 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) pyrimidine nucleoside catabolic process(GO:0046135) |
| 0.1 | 0.7 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) |
| 0.1 | 0.4 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.1 | 2.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.5 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 1.8 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 1.0 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 3.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 4.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 2.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.3 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 0.6 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 4.4 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 0.6 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 1.2 | GO:1902739 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.1 | 0.7 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 1.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.4 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.2 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 | 1.1 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 1.1 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.5 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.9 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.6 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 1.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.4 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.5 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 3.9 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 7.7 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 0.9 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 2.4 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 0.4 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.1 | 0.2 | GO:0046121 | dGTP metabolic process(GO:0046070) deoxyribonucleoside catabolic process(GO:0046121) purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 0.6 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.9 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 0.8 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 2.8 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 1.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.6 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 2.1 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.1 | 2.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 1.0 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.1 | 2.0 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 2.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 0.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 1.1 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.8 | GO:0071236 | cellular response to antibiotic(GO:0071236) |
| 0.1 | 1.1 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 0.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.6 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 1.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 1.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.4 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.9 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.7 | GO:0051324 | meiotic prophase I(GO:0007128) prophase(GO:0051324) |
| 0.1 | 1.8 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 3.4 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 0.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 2.6 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 0.8 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 2.0 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.1 | 0.8 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.3 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.7 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 0.2 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 7.9 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 1.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.3 | GO:0001806 | type IV hypersensitivity(GO:0001806) |
| 0.1 | 1.3 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 | 0.7 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.1 | 0.5 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.1 | 0.9 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.1 | 1.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.5 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.4 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 1.7 | GO:0045187 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 1.3 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.6 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 4.4 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.1 | 0.3 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.4 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.1 | 0.7 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.1 | 0.6 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 1.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.8 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 0.2 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 1.6 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 5.6 | GO:0048709 | oligodendrocyte differentiation(GO:0048709) |
| 0.1 | 2.9 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 0.4 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.1 | 0.6 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.3 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 0.7 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.1 | 1.2 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 0.2 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 0.4 | GO:1902548 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.1 | 0.3 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.3 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 1.5 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 0.7 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 1.8 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.5 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.1 | 0.4 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 | 1.1 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 1.4 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.2 | GO:1900227 | alpha-tubulin acetylation(GO:0071929) positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 2.9 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 1.4 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.6 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.1 | 1.3 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.1 | GO:0071873 | epinephrine metabolic process(GO:0042414) response to norepinephrine(GO:0071873) |
| 0.0 | 0.3 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 3.7 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.4 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.0 | 0.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 3.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.5 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 1.1 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 1.3 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 2.2 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 0.8 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.3 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.8 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.2 | GO:0016093 | polyprenol metabolic process(GO:0016093) |
| 0.0 | 0.7 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.6 | GO:0044036 | cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 2.0 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.6 | GO:0044550 | secondary metabolite biosynthetic process(GO:0044550) |
| 0.0 | 0.3 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 1.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.2 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.0 | 0.2 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.7 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.5 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.3 | GO:0001659 | temperature homeostasis(GO:0001659) |
| 0.0 | 0.1 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.2 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.4 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.6 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.5 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.9 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.3 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 1.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 1.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.7 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.2 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.9 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.0 | 0.0 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.5 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 5.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.5 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 1.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.7 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.8 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 2.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.8 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.4 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 1.0 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 2.0 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 1.0 | GO:0001707 | mesoderm formation(GO:0001707) |
| 0.0 | 1.0 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 0.9 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.0 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.4 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.3 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 2.2 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 0.6 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 0.1 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.2 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.0 | 0.4 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.1 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.8 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 2.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.3 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.0 | 0.2 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.9 | GO:0036503 | ERAD pathway(GO:0036503) |
| 0.0 | 0.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.9 | GO:0034341 | response to interferon-gamma(GO:0034341) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 | 1.2 | GO:0008033 | tRNA processing(GO:0008033) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 15.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.3 | 5.4 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 1.3 | 3.9 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.7 | 2.7 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.6 | 7.3 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.5 | 1.5 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.5 | 1.5 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.5 | 2.6 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.5 | 2.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.5 | 6.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.5 | 2.8 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.4 | 1.7 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.4 | 1.1 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
| 0.4 | 1.5 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.3 | 2.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.3 | 4.5 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.3 | 2.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 1.6 | GO:0072487 | MSL complex(GO:0072487) |
| 0.3 | 1.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 0.9 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.3 | 0.8 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.3 | 1.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.3 | 3.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 1.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 1.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 22.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.2 | 3.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.2 | 0.7 | GO:0044299 | C-fiber(GO:0044299) |
| 0.2 | 2.9 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 1.6 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 2.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 31.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 1.9 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 1.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 0.6 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.2 | 1.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 1.0 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.2 | 4.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 1.1 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 0.4 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.2 | 2.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 2.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 2.1 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.2 | 1.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 2.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 0.5 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.2 | 0.5 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.2 | 2.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 0.9 | GO:0071547 | piP-body(GO:0071547) |
| 0.2 | 3.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 4.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 1.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 1.2 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 2.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.9 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 2.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 2.0 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 3.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 2.9 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 2.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 12.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.6 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 1.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 1.0 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 3.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 2.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 3.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.7 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 1.2 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.1 | GO:0071920 | cleavage body(GO:0071920) |
| 0.1 | 0.8 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 5.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.5 | GO:0036396 | MIS complex(GO:0036396) |
| 0.1 | 0.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.7 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 0.7 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 3.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.4 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 30.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 1.6 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 0.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.3 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 2.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.6 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.8 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 1.0 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 1.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.4 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 2.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 1.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.6 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 2.2 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 2.5 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 0.3 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.5 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 2.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 2.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.5 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 8.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 3.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 18.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 1.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.3 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 5.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 1.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 57.4 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.0 | GO:0044298 | cell body membrane(GO:0044298) |
| 0.0 | 3.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 2.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 3.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 1.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 13.2 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 2.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.2 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 14.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.5 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.6 | 43.2 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 7.2 | 21.6 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 5.0 | 14.9 | GO:0005186 | pheromone activity(GO:0005186) |
| 3.3 | 13.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 2.8 | 11.4 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 2.6 | 10.4 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 2.2 | 6.6 | GO:0008431 | vitamin E binding(GO:0008431) |
| 2.1 | 6.3 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 1.9 | 5.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.8 | 5.3 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 1.7 | 10.4 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 1.7 | 8.6 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 1.5 | 22.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 1.5 | 4.4 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 1.4 | 5.7 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 1.1 | 4.5 | GO:1904121 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 1.1 | 5.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 1.1 | 3.2 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 1.0 | 3.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 1.0 | 3.9 | GO:0004058 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 1.0 | 4.8 | GO:0032810 | sterol response element binding(GO:0032810) |
| 1.0 | 21.9 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.9 | 9.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.8 | 3.2 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.8 | 4.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.8 | 2.4 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.8 | 8.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.7 | 3.7 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.7 | 2.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.6 | 1.9 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.6 | 1.9 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.6 | 2.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.6 | 2.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.6 | 8.9 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.6 | 8.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.6 | 21.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.6 | 2.3 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.6 | 3.3 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.5 | 2.7 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.5 | 3.8 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.5 | 1.6 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.5 | 15.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.5 | 1.6 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.5 | 1.6 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.5 | 1.5 | GO:0004794 | L-serine ammonia-lyase activity(GO:0003941) L-threonine ammonia-lyase activity(GO:0004794) |
| 0.5 | 2.5 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.5 | 1.5 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.5 | 6.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.5 | 1.4 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.4 | 3.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.4 | 4.9 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.4 | 1.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.4 | 3.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.4 | 1.3 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.4 | 3.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.4 | 3.8 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.4 | 2.5 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.4 | 1.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.4 | 2.5 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.4 | 2.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.4 | 2.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.4 | 1.6 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.4 | 1.2 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.4 | 2.3 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.4 | 2.0 | GO:0052796 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.4 | 2.3 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.4 | 1.1 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.4 | 3.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.4 | 8.9 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.4 | 1.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.4 | 7.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 1.4 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.3 | 1.7 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.3 | 1.4 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.3 | 2.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.3 | 1.7 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.3 | 2.6 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.3 | 1.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.3 | 3.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.3 | 1.5 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.3 | 1.8 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.3 | 0.9 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.3 | 1.8 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.3 | 1.4 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.3 | 1.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.3 | 3.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.3 | 2.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 0.8 | GO:0030977 | taurine binding(GO:0030977) |
| 0.3 | 2.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 1.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.3 | 3.7 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.3 | 6.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 2.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.3 | 2.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.2 | 2.9 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 10.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 2.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 0.7 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.2 | 45.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 4.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 0.7 | GO:0030617 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) |
| 0.2 | 0.7 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.2 | 3.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 1.9 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 0.6 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.2 | 1.5 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 1.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.2 | 0.8 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 3.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 0.6 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.2 | 1.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.2 | 1.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 1.5 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 1.3 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.2 | 6.7 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.2 | 2.7 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.2 | 0.8 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 1.5 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.2 | 3.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 2.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 3.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 1.4 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.2 | 0.9 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 1.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 3.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.2 | 0.7 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.2 | 0.9 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 0.7 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.2 | 0.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 2.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 1.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 2.3 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) |
| 0.2 | 0.6 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.2 | 21.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 3.0 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 0.6 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.2 | 0.9 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.2 | 0.5 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.2 | 2.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 2.7 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 0.6 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 1.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.7 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.1 | 0.9 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 3.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 1.7 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 3.2 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.7 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.1 | 2.8 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.0 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 3.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 2.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.4 | GO:0071820 | N-box binding(GO:0071820) |
| 0.1 | 2.0 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 1.1 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 1.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.5 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.5 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.5 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 2.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.9 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 1.4 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.1 | 4.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.3 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.1 | 0.7 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 2.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 2.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 0.4 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 1.0 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 0.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 2.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.7 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.1 | 0.8 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.1 | 1.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.5 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 2.0 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.7 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.4 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 1.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.9 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 4.0 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 0.7 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.5 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 2.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.5 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 0.6 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.6 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.2 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.4 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.3 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 2.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 1.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.6 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 3.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 1.3 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.1 | 2.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.6 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.1 | 2.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 2.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 0.2 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.1 | 0.7 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 2.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.5 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 1.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.3 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.9 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 4.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 5.9 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 1.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 1.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 2.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 1.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 2.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.0 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 1.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 2.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 4.9 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.7 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 2.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 1.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.0 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.1 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 1.3 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) phosphofructokinase activity(GO:0008443) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 1.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 4.1 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 1.3 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 1.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 2.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.8 | GO:0017171 | serine-type peptidase activity(GO:0008236) serine hydrolase activity(GO:0017171) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.9 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 1.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.8 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.4 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.6 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 12.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 7.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 6.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 10.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 4.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 6.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 0.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 4.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 5.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 3.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 0.7 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 1.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 3.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 2.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 2.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 3.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.3 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 1.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 1.5 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 2.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.5 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 1.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 26.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 1.3 | 11.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.7 | 2.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.6 | 7.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.5 | 5.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.5 | 1.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.5 | 4.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.4 | 4.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 13.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.4 | 5.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.4 | 2.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.4 | 6.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.4 | 3.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.3 | 4.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 0.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.3 | 4.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 10.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.3 | 3.0 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 4.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 13.9 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.2 | 0.7 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.2 | 4.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 2.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 4.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 3.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 3.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 9.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.2 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 1.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 3.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.3 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.1 | 1.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 4.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.8 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 9.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 3.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 2.0 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 8.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 3.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 4.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 3.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 2.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 1.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 1.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 0.9 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 2.2 | REACTOME SHC MEDIATED CASCADE | Genes involved in SHC-mediated cascade |
| 0.1 | 2.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.1 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 0.9 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 1.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 2.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 4.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 2.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.7 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.5 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |