Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
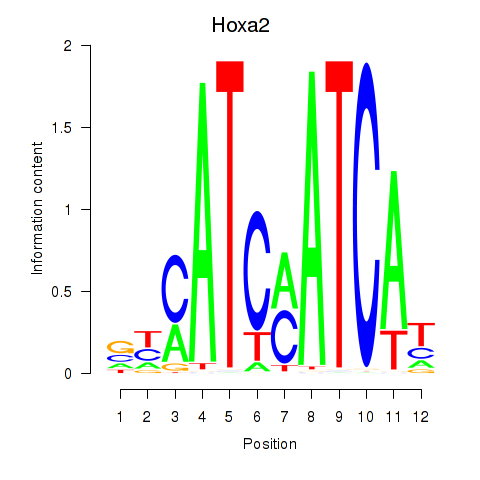
Results for Hoxa2
Z-value: 1.70
Transcription factors associated with Hoxa2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa2
|
ENSMUSG00000014704.8 | homeobox A2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa2 | mm10_v2_chr6_-_52165413_52165413 | -0.47 | 3.7e-03 | Click! |
Activity profile of Hoxa2 motif
Sorted Z-values of Hoxa2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_63344548 | 29.77 |
ENSMUST00000030044.2
|
Orm1
|
orosomucoid 1 |
| chr4_-_62054112 | 25.44 |
ENSMUST00000074018.3
|
Mup20
|
major urinary protein 20 |
| chr4_-_62150810 | 15.94 |
ENSMUST00000077719.3
|
Mup21
|
major urinary protein 21 |
| chr19_-_8131982 | 14.92 |
ENSMUST00000065651.4
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr19_-_39463067 | 12.06 |
ENSMUST00000035488.2
|
Cyp2c38
|
cytochrome P450, family 2, subfamily c, polypeptide 38 |
| chr7_-_46742979 | 11.99 |
ENSMUST00000128088.1
|
Saa1
|
serum amyloid A 1 |
| chr7_+_27119909 | 11.23 |
ENSMUST00000003100.8
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr2_+_102706356 | 10.69 |
ENSMUST00000123759.1
ENSMUST00000111212.1 ENSMUST00000005220.4 |
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr7_+_44207307 | 10.00 |
ENSMUST00000077354.4
|
Klk1b4
|
kallikrein 1-related pepidase b4 |
| chr5_-_87254804 | 9.93 |
ENSMUST00000075858.3
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr12_-_103457195 | 9.86 |
ENSMUST00000044687.6
|
Ifi27l2b
|
interferon, alpha-inducible protein 27 like 2B |
| chr9_-_48605147 | 9.79 |
ENSMUST00000034808.5
ENSMUST00000119426.1 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr7_+_46751832 | 9.74 |
ENSMUST00000075982.2
|
Saa2
|
serum amyloid A 2 |
| chr2_+_25700039 | 9.65 |
ENSMUST00000077667.3
|
Obp2a
|
odorant binding protein 2A |
| chr5_-_146009598 | 9.61 |
ENSMUST00000138870.1
ENSMUST00000068317.6 |
Cyp3a25
|
cytochrome P450, family 3, subfamily a, polypeptide 25 |
| chr4_-_108118504 | 9.01 |
ENSMUST00000149106.1
|
Scp2
|
sterol carrier protein 2, liver |
| chr19_+_4711153 | 8.50 |
ENSMUST00000008991.6
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr12_-_104013640 | 8.10 |
ENSMUST00000058464.4
|
Serpina9
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 |
| chr19_+_12633507 | 7.91 |
ENSMUST00000119960.1
|
Glyat
|
glycine-N-acyltransferase |
| chr8_-_5105232 | 7.83 |
ENSMUST00000023835.1
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr4_+_63356152 | 7.80 |
ENSMUST00000006687.4
|
Orm3
|
orosomucoid 3 |
| chr19_-_4042165 | 7.03 |
ENSMUST00000042700.9
|
Gstp2
|
glutathione S-transferase, pi 2 |
| chr11_-_58613481 | 6.86 |
ENSMUST00000048801.7
|
2210407C18Rik
|
RIKEN cDNA 2210407C18 gene |
| chr3_+_130617645 | 6.86 |
ENSMUST00000163620.1
|
Etnppl
|
ethanolamine phosphate phospholyase |
| chr3_+_130617448 | 6.72 |
ENSMUST00000166187.1
ENSMUST00000072271.6 |
Etnppl
|
ethanolamine phosphate phospholyase |
| chr1_-_121327734 | 6.11 |
ENSMUST00000160968.1
ENSMUST00000162582.1 |
Insig2
|
insulin induced gene 2 |
| chr18_-_61911783 | 5.93 |
ENSMUST00000049378.8
ENSMUST00000166783.1 |
Ablim3
|
actin binding LIM protein family, member 3 |
| chr19_+_12633303 | 5.84 |
ENSMUST00000044976.5
|
Glyat
|
glycine-N-acyltransferase |
| chr5_-_87424201 | 5.82 |
ENSMUST00000072818.5
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr14_+_65971049 | 5.81 |
ENSMUST00000128539.1
|
Clu
|
clusterin |
| chr15_+_82555087 | 5.64 |
ENSMUST00000068861.6
|
Cyp2d12
|
cytochrome P450, family 2, subfamily d, polypeptide 12 |
| chr12_+_108334341 | 5.54 |
ENSMUST00000021684.4
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr5_-_66173051 | 5.40 |
ENSMUST00000113726.1
|
Rbm47
|
RNA binding motif protein 47 |
| chr5_-_87140318 | 5.38 |
ENSMUST00000067790.6
ENSMUST00000113327.1 |
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr3_-_107943362 | 5.35 |
ENSMUST00000106683.1
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr4_-_108118528 | 5.32 |
ENSMUST00000030340.8
|
Scp2
|
sterol carrier protein 2, liver |
| chrX_-_143933089 | 5.32 |
ENSMUST00000087313.3
|
Dcx
|
doublecortin |
| chr15_+_82452372 | 5.25 |
ENSMUST00000089129.5
|
Cyp2d9
|
cytochrome P450, family 2, subfamily d, polypeptide 9 |
| chr8_-_110039330 | 5.24 |
ENSMUST00000109222.2
|
Chst4
|
carbohydrate (chondroitin 6/keratan) sulfotransferase 4 |
| chr1_+_88134786 | 5.18 |
ENSMUST00000113134.1
ENSMUST00000140092.1 |
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr1_-_139858684 | 5.17 |
ENSMUST00000094489.3
|
Cfhr2
|
complement factor H-related 2 |
| chr6_-_83677807 | 5.12 |
ENSMUST00000037882.6
|
Cd207
|
CD207 antigen |
| chr17_+_3397189 | 5.10 |
ENSMUST00000072156.6
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr2_-_84775420 | 5.03 |
ENSMUST00000111641.1
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr3_-_107943705 | 4.97 |
ENSMUST00000106680.1
ENSMUST00000106684.1 ENSMUST00000106685.2 |
Gstm6
|
glutathione S-transferase, mu 6 |
| chr16_+_96361654 | 4.92 |
ENSMUST00000113794.1
|
Igsf5
|
immunoglobulin superfamily, member 5 |
| chr8_-_94696223 | 4.79 |
ENSMUST00000034227.4
|
Pllp
|
plasma membrane proteolipid |
| chr14_+_30886521 | 4.73 |
ENSMUST00000168782.1
|
Itih4
|
inter alpha-trypsin inhibitor, heavy chain 4 |
| chr14_+_30886476 | 4.69 |
ENSMUST00000006703.6
ENSMUST00000078490.5 ENSMUST00000120269.2 |
Itih4
|
inter alpha-trypsin inhibitor, heavy chain 4 |
| chr2_-_84775388 | 4.67 |
ENSMUST00000023994.3
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr1_-_121327672 | 4.64 |
ENSMUST00000159085.1
ENSMUST00000159125.1 ENSMUST00000161818.1 |
Insig2
|
insulin induced gene 2 |
| chr17_-_34804546 | 4.60 |
ENSMUST00000025223.8
|
Cyp21a1
|
cytochrome P450, family 21, subfamily a, polypeptide 1 |
| chr1_+_88103229 | 4.56 |
ENSMUST00000113135.3
ENSMUST00000113138.1 |
Ugt1a6a
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A6A UDP glucuronosyltransferase 1 family, polypeptide A6B |
| chr11_+_83703991 | 4.50 |
ENSMUST00000092836.5
|
Wfdc17
|
WAP four-disulfide core domain 17 |
| chr1_-_139560158 | 4.44 |
ENSMUST00000160423.1
ENSMUST00000023965.5 |
Cfhr1
|
complement factor H-related 1 |
| chr13_+_4059565 | 4.42 |
ENSMUST00000041768.6
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr9_+_22225702 | 4.38 |
ENSMUST00000072465.7
|
Zfp809
|
zinc finger protein 809 |
| chr10_-_41611319 | 4.34 |
ENSMUST00000179614.1
|
Ccdc162
|
coiled-coil domain containing 162 |
| chr6_-_89216237 | 4.31 |
ENSMUST00000079186.2
|
Gm839
|
predicted gene 839 |
| chr19_+_37685581 | 4.28 |
ENSMUST00000073391.4
|
Cyp26c1
|
cytochrome P450, family 26, subfamily c, polypeptide 1 |
| chr1_-_121327776 | 4.26 |
ENSMUST00000160688.1
|
Insig2
|
insulin induced gene 2 |
| chr6_+_124304646 | 4.20 |
ENSMUST00000112541.2
ENSMUST00000032234.2 |
Cd163
|
CD163 antigen |
| chr15_-_82620907 | 4.17 |
ENSMUST00000109515.1
|
Cyp2d34
|
cytochrome P450, family 2, subfamily d, polypeptide 34 |
| chr13_+_23807027 | 4.17 |
ENSMUST00000006786.4
ENSMUST00000099697.2 |
Slc17a2
|
solute carrier family 17 (sodium phosphate), member 2 |
| chr11_+_102041509 | 4.13 |
ENSMUST00000123895.1
ENSMUST00000017453.5 ENSMUST00000107163.2 ENSMUST00000107164.2 |
Cd300lg
|
CD300 antigen like family member G |
| chr9_-_70141484 | 4.13 |
ENSMUST00000034749.8
|
Fam81a
|
family with sequence similarity 81, member A |
| chr11_-_106580594 | 4.12 |
ENSMUST00000153870.1
|
Tex2
|
testis expressed gene 2 |
| chr3_-_107943390 | 4.10 |
ENSMUST00000106681.1
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr3_+_62419668 | 4.05 |
ENSMUST00000161057.1
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr14_+_51091077 | 4.04 |
ENSMUST00000022428.5
ENSMUST00000171688.1 |
Rnase4
Ang
|
ribonuclease, RNase A family 4 angiogenin, ribonuclease, RNase A family, 5 |
| chr7_+_24587543 | 3.89 |
ENSMUST00000077191.6
|
Ethe1
|
ethylmalonic encephalopathy 1 |
| chr1_-_153900198 | 3.82 |
ENSMUST00000123490.1
|
5830403L16Rik
|
RIKEN cDNA 5830403L16 gene |
| chr16_+_43247278 | 3.82 |
ENSMUST00000114691.1
ENSMUST00000079441.6 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr14_+_65971164 | 3.81 |
ENSMUST00000144619.1
|
Clu
|
clusterin |
| chr3_+_81996922 | 3.75 |
ENSMUST00000029641.3
|
Asic5
|
acid-sensing (proton-gated) ion channel family member 5 |
| chr7_-_100658364 | 3.72 |
ENSMUST00000107043.1
|
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr6_-_28126125 | 3.69 |
ENSMUST00000115324.2
ENSMUST00000090512.3 |
Grm8
|
glutamate receptor, metabotropic 8 |
| chr15_+_31224371 | 3.69 |
ENSMUST00000044524.9
|
Dap
|
death-associated protein |
| chr6_+_121346618 | 3.66 |
ENSMUST00000032200.9
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr13_-_4279420 | 3.61 |
ENSMUST00000021632.3
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr1_+_153899937 | 3.60 |
ENSMUST00000086199.5
|
Glul
|
glutamate-ammonia ligase (glutamine synthetase) |
| chr1_+_191978247 | 3.58 |
ENSMUST00000175680.1
|
Rd3
|
retinal degeneration 3 |
| chr18_-_56562261 | 3.56 |
ENSMUST00000066208.6
ENSMUST00000172734.1 |
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr7_-_71351485 | 3.52 |
ENSMUST00000094315.2
|
Gm10295
|
predicted gene 10295 |
| chr6_-_21851914 | 3.47 |
ENSMUST00000134635.1
ENSMUST00000123116.1 ENSMUST00000120965.1 |
Tspan12
|
tetraspanin 12 |
| chr4_-_109202217 | 3.38 |
ENSMUST00000160774.1
ENSMUST00000030288.7 ENSMUST00000162787.2 |
Osbpl9
|
oxysterol binding protein-like 9 |
| chr18_-_56562187 | 3.38 |
ENSMUST00000171844.2
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr5_-_130024280 | 3.34 |
ENSMUST00000161640.1
ENSMUST00000161884.1 ENSMUST00000161094.1 |
Asl
|
argininosuccinate lyase |
| chr2_-_91195035 | 3.27 |
ENSMUST00000111356.1
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr7_-_98562982 | 3.24 |
ENSMUST00000069035.4
|
A630091E08Rik
|
RIKEN cDNA A630091E08 gene |
| chr3_-_107931817 | 3.19 |
ENSMUST00000004137.4
|
Gstm7
|
glutathione S-transferase, mu 7 |
| chr4_-_96507386 | 3.18 |
ENSMUST00000124729.3
|
Cyp2j8
|
cytochrome P450, family 2, subfamily j, polypeptide 8 |
| chr13_+_4233730 | 3.18 |
ENSMUST00000081326.6
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr18_-_56562215 | 3.15 |
ENSMUST00000170309.1
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr13_-_41847626 | 3.00 |
ENSMUST00000121404.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr3_-_113574758 | 2.98 |
ENSMUST00000106540.1
|
Amy1
|
amylase 1, salivary |
| chr13_+_4049001 | 2.97 |
ENSMUST00000118717.2
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr3_-_157925056 | 2.96 |
ENSMUST00000118539.1
|
Cth
|
cystathionase (cystathionine gamma-lyase) |
| chr10_-_18227473 | 2.94 |
ENSMUST00000174592.1
|
Ccdc28a
|
coiled-coil domain containing 28A |
| chr13_-_41847599 | 2.93 |
ENSMUST00000179758.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr15_+_99717515 | 2.88 |
ENSMUST00000023760.6
ENSMUST00000162194.1 |
Gpd1
|
glycerol-3-phosphate dehydrogenase 1 (soluble) |
| chr15_+_31568791 | 2.87 |
ENSMUST00000162532.1
|
Cmbl
|
carboxymethylenebutenolidase-like (Pseudomonas) |
| chr7_-_24587612 | 2.85 |
ENSMUST00000094705.2
|
Zfp575
|
zinc finger protein 575 |
| chr4_-_115496129 | 2.81 |
ENSMUST00000030487.2
|
Cyp4a14
|
cytochrome P450, family 4, subfamily a, polypeptide 14 |
| chr11_+_104231390 | 2.80 |
ENSMUST00000106992.3
|
Mapt
|
microtubule-associated protein tau |
| chr3_-_39359128 | 2.73 |
ENSMUST00000056409.2
|
Gm9845
|
predicted pseudogene 9845 |
| chr18_-_3299537 | 2.70 |
ENSMUST00000129435.1
ENSMUST00000122958.1 |
Crem
|
cAMP responsive element modulator |
| chr7_+_143823135 | 2.67 |
ENSMUST00000128454.1
ENSMUST00000073878.5 |
Dhcr7
|
7-dehydrocholesterol reductase |
| chr10_-_75797528 | 2.63 |
ENSMUST00000120177.1
|
Gstt1
|
glutathione S-transferase, theta 1 |
| chrX_-_19237841 | 2.61 |
ENSMUST00000180592.1
|
Gm26652
|
predicted gene, 26652 |
| chr3_-_98588807 | 2.59 |
ENSMUST00000178221.1
|
Gm10681
|
predicted gene 10681 |
| chr1_-_90967667 | 2.58 |
ENSMUST00000131428.1
|
Rab17
|
RAB17, member RAS oncogene family |
| chrX_+_7722267 | 2.57 |
ENSMUST00000125991.1
ENSMUST00000148624.1 |
Wdr45
|
WD repeat domain 45 |
| chr10_+_81575257 | 2.57 |
ENSMUST00000135211.1
|
Tle2
|
transducin-like enhancer of split 2, homolog of Drosophila E(spl) |
| chr4_+_95557494 | 2.57 |
ENSMUST00000079223.4
ENSMUST00000177394.1 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr6_+_48554776 | 2.56 |
ENSMUST00000114545.1
ENSMUST00000153222.1 ENSMUST00000101436.2 |
Lrrc61
|
leucine rich repeat containing 61 |
| chrX_+_7722214 | 2.56 |
ENSMUST00000043045.2
ENSMUST00000116634.1 ENSMUST00000115689.3 ENSMUST00000131077.1 ENSMUST00000115688.1 ENSMUST00000116633.1 |
Wdr45
|
WD repeat domain 45 |
| chr6_+_108828633 | 2.53 |
ENSMUST00000089162.3
|
Edem1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr10_+_81575306 | 2.51 |
ENSMUST00000146916.1
|
Tle2
|
transducin-like enhancer of split 2, homolog of Drosophila E(spl) |
| chr10_-_109009055 | 2.51 |
ENSMUST00000156979.1
|
Syt1
|
synaptotagmin I |
| chrX_-_143933204 | 2.49 |
ENSMUST00000112851.1
ENSMUST00000112856.2 ENSMUST00000033642.3 |
Dcx
|
doublecortin |
| chr4_-_83285141 | 2.48 |
ENSMUST00000150522.1
|
Ttc39b
|
tetratricopeptide repeat domain 39B |
| chr12_+_9029982 | 2.45 |
ENSMUST00000085741.1
|
Ttc32
|
tetratricopeptide repeat domain 32 |
| chr17_-_31165045 | 2.45 |
ENSMUST00000024831.6
|
Tff1
|
trefoil factor 1 |
| chr18_+_9707639 | 2.45 |
ENSMUST00000040069.8
|
Colec12
|
collectin sub-family member 12 |
| chr7_+_43995833 | 2.44 |
ENSMUST00000007156.4
|
Klk1b11
|
kallikrein 1-related peptidase b11 |
| chr9_+_75037614 | 2.43 |
ENSMUST00000168166.1
ENSMUST00000169492.1 ENSMUST00000170308.1 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr2_-_134554348 | 2.39 |
ENSMUST00000028704.2
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr15_+_59648350 | 2.38 |
ENSMUST00000067543.6
|
Trib1
|
tribbles homolog 1 (Drosophila) |
| chr1_+_88055377 | 2.36 |
ENSMUST00000138182.1
ENSMUST00000113142.3 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr7_+_140125651 | 2.35 |
ENSMUST00000026537.5
ENSMUST00000097967.3 |
Paox
|
polyamine oxidase (exo-N4-amino) |
| chr7_+_113207465 | 2.33 |
ENSMUST00000047321.7
|
Arntl
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr9_+_123021315 | 2.29 |
ENSMUST00000084733.5
|
Tmem42
|
transmembrane protein 42 |
| chr19_+_24875679 | 2.26 |
ENSMUST00000073080.5
|
Gm10053
|
predicted gene 10053 |
| chr17_+_37193889 | 2.26 |
ENSMUST00000038844.6
|
Ubd
|
ubiquitin D |
| chr9_+_75037744 | 2.25 |
ENSMUST00000168301.1
|
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr7_-_46715676 | 2.25 |
ENSMUST00000006956.7
|
Saa3
|
serum amyloid A 3 |
| chr12_+_57564111 | 2.24 |
ENSMUST00000101398.3
|
Ttc6
|
tetratricopeptide repeat domain 6 |
| chr2_+_116900152 | 2.21 |
ENSMUST00000126467.1
ENSMUST00000128305.1 ENSMUST00000155323.1 |
D330050G23Rik
|
RIKEN cDNA D330050G23 gene |
| chr7_-_108930151 | 2.20 |
ENSMUST00000055745.3
|
Nlrp10
|
NLR family, pyrin domain containing 10 |
| chr2_-_91195097 | 2.19 |
ENSMUST00000002177.2
ENSMUST00000111354.1 |
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr13_-_23934156 | 2.18 |
ENSMUST00000052776.2
|
Hist1h2ba
|
histone cluster 1, H2ba |
| chr17_-_32424139 | 2.17 |
ENSMUST00000114455.1
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr19_-_44029201 | 2.17 |
ENSMUST00000026211.8
|
Cyp2c44
|
cytochrome P450, family 2, subfamily c, polypeptide 44 |
| chr10_+_57784914 | 2.16 |
ENSMUST00000165013.1
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr13_-_24937585 | 2.16 |
ENSMUST00000037615.6
|
Aldh5a1
|
aldhehyde dehydrogenase family 5, subfamily A1 |
| chr1_+_88055467 | 2.15 |
ENSMUST00000173325.1
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr10_+_29143996 | 2.15 |
ENSMUST00000092629.2
|
Soga3
|
SOGA family member 3 |
| chr16_+_90220742 | 2.14 |
ENSMUST00000023707.9
|
Sod1
|
superoxide dismutase 1, soluble |
| chr16_-_75766758 | 2.13 |
ENSMUST00000114244.1
ENSMUST00000046283.8 |
Hspa13
|
heat shock protein 70 family, member 13 |
| chr5_+_139543889 | 2.11 |
ENSMUST00000174792.1
ENSMUST00000031523.8 |
Uncx
|
UNC homeobox |
| chr5_+_125475814 | 2.10 |
ENSMUST00000031445.3
|
Aacs
|
acetoacetyl-CoA synthetase |
| chr7_+_29238434 | 2.10 |
ENSMUST00000108237.1
|
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr3_-_88762244 | 2.09 |
ENSMUST00000183267.1
|
Syt11
|
synaptotagmin XI |
| chr9_+_121710389 | 2.07 |
ENSMUST00000035113.9
|
Deb1
|
differentially expressed in B16F10 1 |
| chr18_-_12819842 | 2.07 |
ENSMUST00000119043.1
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr17_+_30485511 | 2.06 |
ENSMUST00000064223.1
|
Gm9874
|
predicted gene 9874 |
| chr4_-_35157404 | 2.06 |
ENSMUST00000102975.3
|
Mob3b
|
MOB kinase activator 3B |
| chr16_-_36990449 | 2.03 |
ENSMUST00000075869.6
|
Fbxo40
|
F-box protein 40 |
| chr9_+_75037809 | 2.00 |
ENSMUST00000167885.1
|
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr11_-_86671169 | 2.00 |
ENSMUST00000143991.1
|
Vmp1
|
vacuole membrane protein 1 |
| chr12_+_4769375 | 2.00 |
ENSMUST00000178879.1
|
Pfn4
|
profilin family, member 4 |
| chr4_+_116596672 | 1.98 |
ENSMUST00000051869.7
|
Ccdc17
|
coiled-coil domain containing 17 |
| chrX_+_100428906 | 1.98 |
ENSMUST00000060241.2
|
Otud6a
|
OTU domain containing 6A |
| chr9_+_75037712 | 1.98 |
ENSMUST00000007800.7
|
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr8_+_47824459 | 1.97 |
ENSMUST00000038693.6
|
Cldn22
|
claudin 22 |
| chr5_+_3596066 | 1.96 |
ENSMUST00000006061.6
ENSMUST00000121291.1 ENSMUST00000142516.1 |
Pex1
|
peroxisomal biogenesis factor 1 |
| chr2_-_177925604 | 1.92 |
ENSMUST00000108934.2
ENSMUST00000081529.4 |
C330013J21Rik
|
RIKEN cDNA C330013J21 gene |
| chr11_-_40755201 | 1.92 |
ENSMUST00000020576.7
|
Ccng1
|
cyclin G1 |
| chr6_-_142278836 | 1.91 |
ENSMUST00000111825.3
|
Slco1a5
|
solute carrier organic anion transporter family, member 1a5 |
| chr18_-_35498856 | 1.91 |
ENSMUST00000025215.8
|
Sil1
|
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
| chr4_+_102589687 | 1.90 |
ENSMUST00000097949.4
ENSMUST00000106901.1 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr7_+_130936172 | 1.90 |
ENSMUST00000006367.7
|
Htra1
|
HtrA serine peptidase 1 |
| chr2_+_96318014 | 1.90 |
ENSMUST00000135431.1
ENSMUST00000162807.2 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr14_+_51007911 | 1.88 |
ENSMUST00000022424.6
|
Rnase10
|
ribonuclease, RNase A family, 10 (non-active) |
| chr15_+_59315030 | 1.85 |
ENSMUST00000022977.7
|
Sqle
|
squalene epoxidase |
| chr8_+_21938352 | 1.84 |
ENSMUST00000076786.2
|
Defb35
|
defensin beta 35 |
| chr10_+_81575499 | 1.84 |
ENSMUST00000143285.1
ENSMUST00000146358.1 |
Tle2
|
transducin-like enhancer of split 2, homolog of Drosophila E(spl) |
| chr10_+_57784859 | 1.84 |
ENSMUST00000020024.5
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr4_-_147936713 | 1.83 |
ENSMUST00000105712.1
ENSMUST00000019199.7 |
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chrX_+_142226765 | 1.82 |
ENSMUST00000112916.2
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr9_+_123806468 | 1.80 |
ENSMUST00000049810.7
|
Cxcr6
|
chemokine (C-X-C motif) receptor 6 |
| chr19_-_8218832 | 1.77 |
ENSMUST00000113298.2
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr9_+_75037838 | 1.76 |
ENSMUST00000169188.1
|
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr18_-_3299452 | 1.73 |
ENSMUST00000126578.1
|
Crem
|
cAMP responsive element modulator |
| chr11_+_94327984 | 1.73 |
ENSMUST00000107818.2
ENSMUST00000051221.6 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr1_+_107422681 | 1.72 |
ENSMUST00000112710.1
ENSMUST00000086690.4 |
Serpinb7
|
serine (or cysteine) peptidase inhibitor, clade B, member 7 |
| chr6_-_114969986 | 1.72 |
ENSMUST00000139640.1
|
Vgll4
|
vestigial like 4 (Drosophila) |
| chrX_-_134161928 | 1.72 |
ENSMUST00000033611.4
|
Xkrx
|
X Kell blood group precursor related X linked |
| chr3_-_65392579 | 1.71 |
ENSMUST00000029414.5
|
Ssr3
|
signal sequence receptor, gamma |
| chr1_-_155527083 | 1.71 |
ENSMUST00000097531.2
|
Gm5532
|
predicted gene 5532 |
| chrX_-_36874111 | 1.69 |
ENSMUST00000047486.5
|
C330007P06Rik
|
RIKEN cDNA C330007P06 gene |
| chr13_-_4609122 | 1.67 |
ENSMUST00000110691.3
ENSMUST00000091848.5 |
Akr1e1
|
aldo-keto reductase family 1, member E1 |
| chr6_-_24527546 | 1.67 |
ENSMUST00000118558.1
|
Ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr14_-_56159579 | 1.67 |
ENSMUST00000015578.4
|
Gzmg
|
granzyme G |
| chr18_-_6136057 | 1.65 |
ENSMUST00000182559.1
|
Arhgap12
|
Rho GTPase activating protein 12 |
| chr11_+_104231573 | 1.64 |
ENSMUST00000132977.1
ENSMUST00000132245.1 ENSMUST00000100347.4 |
Mapt
|
microtubule-associated protein tau |
| chr15_+_102977032 | 1.63 |
ENSMUST00000001706.6
|
Hoxc9
|
homeobox C9 |
| chr17_+_24426676 | 1.63 |
ENSMUST00000024946.5
|
Eci1
|
enoyl-Coenzyme A delta isomerase 1 |
| chr2_-_60284292 | 1.63 |
ENSMUST00000028356.8
ENSMUST00000074606.4 |
Cd302
|
CD302 antigen |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 25.4 | GO:0008355 | olfactory learning(GO:0008355) |
| 3.6 | 14.3 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 3.5 | 13.9 | GO:0042196 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 3.2 | 9.7 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 1.8 | 5.5 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 1.7 | 5.2 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 1.6 | 9.6 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.5 | 10.7 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.5 | 4.4 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 1.4 | 6.9 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 1.3 | 6.7 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 1.2 | 67.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 1.2 | 16.7 | GO:0015747 | urate transport(GO:0015747) |
| 1.2 | 14.3 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 1.1 | 2.2 | GO:0032824 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 1.1 | 4.3 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 1.0 | 4.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.0 | 3.0 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 1.0 | 2.9 | GO:0006127 | glycerophosphate shuttle(GO:0006127) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.9 | 3.7 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.9 | 2.7 | GO:0016132 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.9 | 15.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.9 | 1.7 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.8 | 2.5 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.8 | 7.3 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.8 | 2.4 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.8 | 3.9 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.7 | 2.1 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.7 | 2.0 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.6 | 1.3 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.6 | 4.8 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.6 | 2.4 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.6 | 4.4 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.5 | 5.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.5 | 3.2 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.5 | 1.6 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.5 | 1.5 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.5 | 2.5 | GO:1902714 | regulation of natural killer cell degranulation(GO:0043321) negative regulation of interferon-gamma secretion(GO:1902714) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.5 | 2.3 | GO:0008295 | spermidine biosynthetic process(GO:0008295) putrescine biosynthetic process(GO:0009446) |
| 0.5 | 1.9 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.5 | 2.3 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.5 | 1.8 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.5 | 1.8 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.4 | 14.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.4 | 7.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.4 | 1.7 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.4 | 2.2 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.4 | 0.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.4 | 1.3 | GO:0002591 | positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) peptide antigen transport(GO:0046968) |
| 0.4 | 15.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.4 | 5.0 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.4 | 1.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.4 | 5.5 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.4 | 1.2 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.4 | 1.1 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.4 | 9.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.4 | 2.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.4 | 2.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.3 | 5.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.3 | 3.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.3 | 1.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 10.4 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.3 | 1.6 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.3 | 10.0 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.3 | 8.5 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.3 | 0.9 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.3 | 1.2 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.3 | 1.1 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.3 | 0.8 | GO:1900114 | positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.3 | 1.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.3 | 1.0 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.2 | 1.5 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.2 | 1.5 | GO:0071484 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.2 | 2.5 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.2 | 2.2 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.2 | 0.9 | GO:0009223 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.2 | 2.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 0.9 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 1.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 2.0 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 4.0 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.2 | 1.9 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.2 | 0.6 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.2 | 1.2 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.2 | 2.0 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.2 | 1.6 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.2 | 1.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 0.8 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.2 | 0.8 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.2 | 4.0 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.2 | 0.5 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 1.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 1.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.2 | 0.9 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 | 1.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 1.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.2 | 2.6 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.2 | 0.5 | GO:0090081 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.2 | 0.5 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.2 | 0.5 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.2 | 0.5 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.2 | 0.7 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 2.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 2.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.2 | 1.7 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.2 | 0.5 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.2 | 1.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 1.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 1.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 1.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 1.9 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 1.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.6 | GO:2000864 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 0.4 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.1 | 0.6 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.1 | 2.1 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 0.5 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.1 | 1.2 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 0.4 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 1.6 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.1 | 3.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.8 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.1 | 1.4 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.5 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.1 | 0.6 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 1.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.3 | GO:0070347 | brown fat cell proliferation(GO:0070342) regulation of brown fat cell proliferation(GO:0070347) |
| 0.1 | 0.9 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 2.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.4 | GO:0002587 | negative regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002587) |
| 0.1 | 0.5 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.1 | 1.0 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 0.4 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.7 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 0.8 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.4 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 1.0 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 3.9 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 0.2 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 3.6 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 0.6 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.6 | GO:0060459 | left lung development(GO:0060459) left lung morphogenesis(GO:0060460) |
| 0.1 | 1.1 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.1 | 0.5 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.6 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 12.2 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 1.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.2 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.3 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 6.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 1.0 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.1 | 3.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 8.6 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 1.0 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 0.7 | GO:2000303 | regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 1.1 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.1 | 0.8 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.1 | 1.4 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.1 | 2.1 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.6 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.1 | 0.9 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 1.6 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.6 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.2 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 1.0 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.1 | 0.5 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 1.6 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 1.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.8 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.7 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.1 | 0.2 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) |
| 0.1 | 0.9 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.8 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.2 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 | 2.3 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 1.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.4 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 1.5 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.7 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.5 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.5 | GO:2000484 | positive regulation of interleukin-8 secretion(GO:2000484) |
| 0.0 | 0.9 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.6 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 2.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 1.1 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 4.8 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.2 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.5 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 1.9 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 2.4 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.1 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 2.1 | GO:0007584 | response to nutrient(GO:0007584) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.0 | 0.7 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.8 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 0.6 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 7.0 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.2 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 2.2 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 4.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.9 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 1.6 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.4 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 1.1 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.3 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.9 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.3 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 0.8 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.0 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.2 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.9 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.0 | 1.0 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.1 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.0 | 0.5 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.0 | 1.6 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.4 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 4.0 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 1.3 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.8 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 1.9 | 15.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.3 | 4.0 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 1.3 | 16.9 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 1.2 | 8.5 | GO:0008091 | spectrin(GO:0008091) |
| 1.0 | 12.7 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.8 | 24.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.6 | 2.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 1.7 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.3 | 5.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.3 | 10.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.3 | 1.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.3 | 0.8 | GO:0060473 | cortical granule(GO:0060473) |
| 0.3 | 3.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.3 | 1.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 4.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 1.4 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 2.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 1.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 0.8 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.2 | 1.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 0.8 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.2 | 1.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 0.5 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.2 | 2.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 4.8 | GO:0043218 | compact myelin(GO:0043218) |
| 0.2 | 1.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 1.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 0.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 21.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 2.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 1.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.5 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 2.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 4.8 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 0.9 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.9 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 1.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.6 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.7 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 2.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 2.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.5 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.1 | 1.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.0 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 5.7 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 1.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 2.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.6 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 1.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.4 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 2.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 3.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.6 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 7.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.5 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 2.3 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.0 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 1.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 1.7 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 3.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 2.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 9.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 4.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 41.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 4.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 2.9 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 1.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.7 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.5 | 25.4 | GO:0005186 | pheromone activity(GO:0005186) |
| 4.5 | 13.6 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 3.6 | 14.4 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 3.6 | 14.3 | GO:1904121 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 3.2 | 9.6 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 2.5 | 10.1 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 2.0 | 12.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 1.8 | 10.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.4 | 4.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 1.3 | 4.0 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 1.2 | 7.4 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 1.1 | 7.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.1 | 16.7 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 1.1 | 5.5 | GO:0032810 | sterol response element binding(GO:0032810) |
| 1.0 | 7.3 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 1.0 | 2.9 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.9 | 34.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.9 | 2.7 | GO:0047598 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.9 | 2.6 | GO:0016824 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.9 | 5.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.9 | 2.6 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.8 | 3.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.8 | 2.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.7 | 21.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.6 | 2.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.6 | 3.7 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.6 | 2.4 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.6 | 1.7 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.5 | 2.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.5 | 3.7 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.5 | 24.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.5 | 11.2 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.5 | 12.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.5 | 1.5 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.5 | 3.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.5 | 2.8 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.5 | 1.8 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.4 | 2.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.4 | 4.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.4 | 3.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.4 | 3.8 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.4 | 10.6 | GO:0001848 | complement binding(GO:0001848) |
| 0.4 | 1.2 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.4 | 9.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 4.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 2.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.3 | 2.7 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.3 | 1.6 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.3 | 1.6 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.3 | 1.6 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.3 | 1.2 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.3 | 1.8 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.3 | 1.2 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.3 | 4.1 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.3 | 0.8 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.3 | 0.8 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.3 | 1.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.3 | 1.3 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.2 | 1.9 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 0.7 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.2 | 2.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 1.6 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.2 | 7.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.2 | 5.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 4.8 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.2 | 1.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 1.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 3.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.2 | 0.7 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.2 | 8.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 3.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.2 | 0.5 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 0.5 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.2 | 1.0 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.2 | 3.9 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 1.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 0.7 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 1.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 2.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 5.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 0.6 | GO:0038100 | nodal binding(GO:0038100) |
| 0.2 | 0.8 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 0.6 | GO:0051381 | histamine binding(GO:0051381) |
| 0.1 | 0.6 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 1.0 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.1 | 0.6 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.1 | 1.3 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 12.1 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 12.4 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 5.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 6.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 16.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.5 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.9 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.3 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 0.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 2.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.9 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.4 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 1.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.6 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 4.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.6 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 4.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.7 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 1.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.5 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 1.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.8 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.2 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.1 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.9 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.5 | GO:0048019 | co-receptor binding(GO:0039706) receptor antagonist activity(GO:0048019) |
| 0.1 | 2.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.4 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.6 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.8 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.2 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 2.2 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.2 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 5.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 2.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.7 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 1.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.6 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.7 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 1.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 6.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 1.0 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.3 | GO:0043855 | cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 1.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.1 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.9 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 1.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.3 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.1 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 2.1 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 1.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.4 | 13.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 7.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 5.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 6.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 6.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 2.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 0.9 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 1.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 2.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 6.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 2.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 27.7 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.6 | 9.9 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.5 | 10.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 8.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 7.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 6.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 2.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 5.9 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 4.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 2.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 14.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.2 | 8.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 2.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 4.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 3.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 9.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 1.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 3.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 12.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 15.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 1.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 5.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 2.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 6.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 3.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 7.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 2.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 4.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 3.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 3.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |